# To install a package, run the following ONCE (and only once on your computer)
# install.packages("psych")
library(here) # makes reading data more consistent
library(tidyverse) # for data manipulation and plotting
library(haven) # for importing SPSS/SAS/Stata data
library(lme4) # for multilevel analysis
library(lmerTest) # for testing in small samples
library(splines) # for nonlinear trend
library(modelsummary) # for tables
library(broom.mixed) # for obtaining residualsWeek 6 (R): Diagnostics
In R, there are quite a few diagnostic tools for MLM, although they may not be straightforward to use. In this note, I will show some basic tools.
Load Packages and Import Data
# Read in the data (pay attention to the directory)
hsball <- read_sav(here("data_files", "hsball.sav"))
# Cluster-mean centering
hsball <- hsball %>%
group_by(id) %>% # operate within schools
mutate(ses_cm = mean(ses), # create cluster means (the same as `meanses`)
ses_cmc = ses - ses_cm) %>% # cluster-mean centered
ungroup() # exit the "editing within groups" modeThe discussion will follow chapter 10 of Snijders & Bosker (2012)
Fitting the model
We will first use a model with ses and sector, without contextual effects or random slopes.
m1 <- lmer(mathach ~ sector + ses + (1 | id),
data = hsball)Diagnostics
Does the fixed part contain the right variables?
Cluster means for lower-level variables
One can test whether ses_cm should be included in the model, in addition to the raw level-1 variable. Let’s use likelihood ratio tests for it:
m1_cm <- lmer(mathach ~ sector + ses + ses_cm + (1 | id),
data = hsball
)
confint(m1_cm)># Computing profile confidence intervals ...># 2.5 % 97.5 %
># .sig01 1.3013272 1.769307
># .sigma 5.9849982 6.186295
># (Intercept) 11.7392184 12.517199
># sector 0.6267857 1.822315
># ses 1.9781613 2.404183
># ses_cm 2.3941778 3.896384So it looks like we need ses_cm.
Omitted variables
If there are any potential confounding variables, those should be included as well. Identifying confounding variables requires knowledge of the research question under investigation. One good thing is that if one is interested in effects at a lower level, doing cluster-mean centering or including the cluster mean will rule out potential confounders at a higher level. This is because one is now comparing individuals within the same school (or the same level-2 unit), so any confounders related to school characteristics can be ruled out, as they will be held constant.
Linearity
A quick way to check whether there are any unmodelled nonlinear trends is to use marginal model plots, which show the outcome variable against each predictor. Each plot shows two lines: one shows a nonparametric smoother that does not depend on the model, and the other shows the implied association based on the model. If the two lines are close to each other, then there is no need to include additional curvilinear terms of the predictors; otherwise, one may need to consider adding quadratic or other nonlinear terms.
m2 <- lmer(mathach ~ sector + ses + ses_cm + (1 | id),
data = hsball
)sjPlot::plot_model(m2, type = "slope", show.data = TRUE)># `geom_smooth()` using formula = 'y ~ x'
># `geom_smooth()` using formula = 'y ~ x'># Warning in simpleLoess(y, x, w, span, degree = degree, parametric =
># parametric, : pseudoinverse used at -0.005># Warning in simpleLoess(y, x, w, span, degree = degree, parametric =
># parametric, : neighborhood radius 1.005># Warning in simpleLoess(y, x, w, span, degree = degree, parametric =
># parametric, : reciprocal condition number 4.3494e-30># Warning in simpleLoess(y, x, w, span, degree = degree, parametric =
># parametric, : There are other near singularities as well. 1.01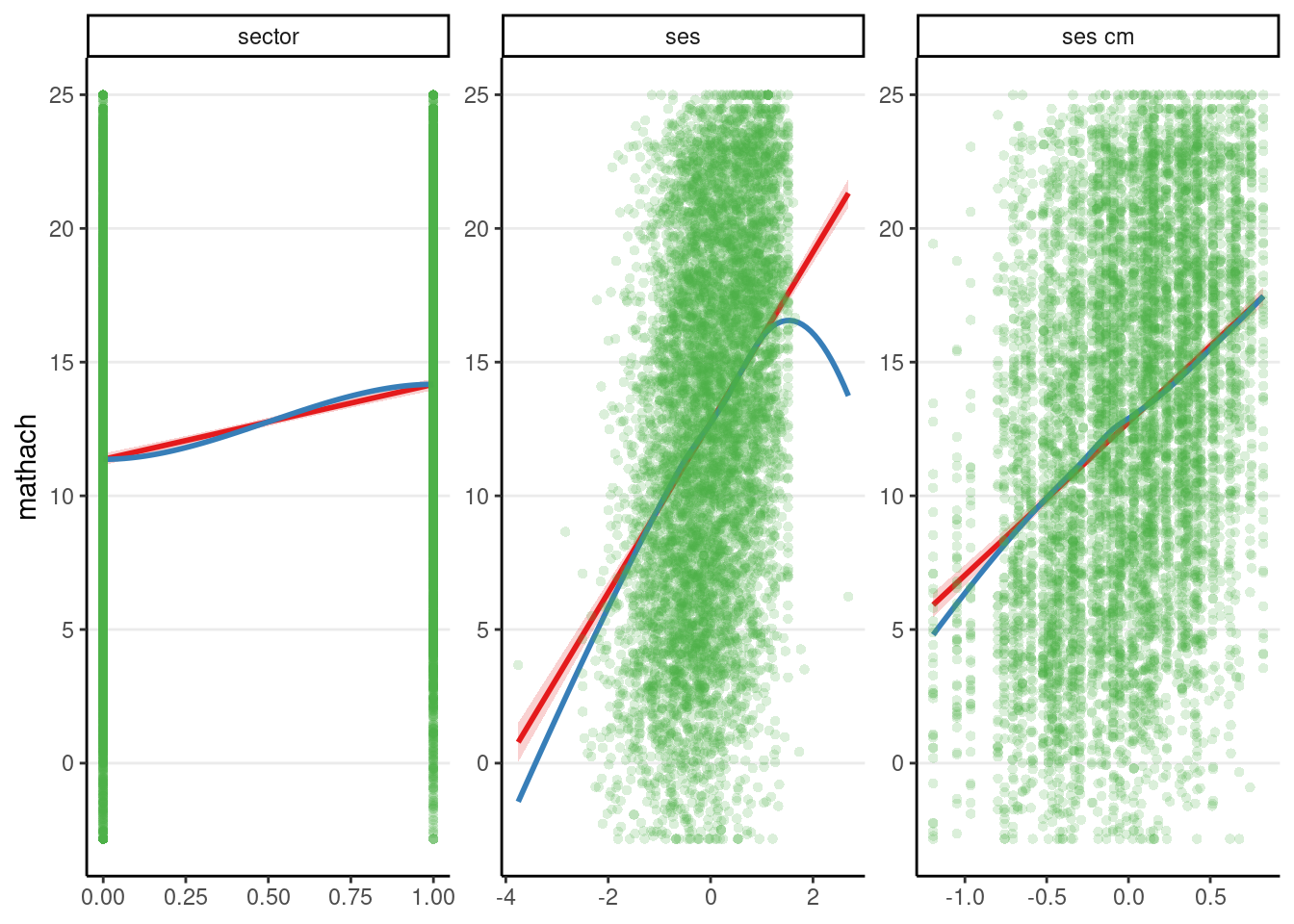
The above shows some deviation between the model (red line) and the data (blue line) for how mathach relates to ses, but a few data points mostly drive the discrepancy, one on the left and one on the right. This is mostly fine, although it is possible to check whether the results are similar without those two points.
For the binary predictor sector, there are only two possible values in the predictor, so the plot may not look as neat, but you can still compare the red and the blue curves.
To show what it is like when linearity is violated, I simulate some data by adding a quadratic term:
## Fake example: Add quadratic trend for ses
hsball_quad <- hsball %>%
mutate(mathach = mathach + ses^2)
m2_quad <- refit(m2, newresp = hsball_quad$mathach)
sjPlot::plot_model(m2_quad, type = "slope", show.data = TRUE)># `geom_smooth()` using formula = 'y ~ x'
># `geom_smooth()` using formula = 'y ~ x'># Warning in simpleLoess(y, x, w, span, degree = degree, parametric =
># parametric, : pseudoinverse used at -0.005># Warning in simpleLoess(y, x, w, span, degree = degree, parametric =
># parametric, : neighborhood radius 1.005># Warning in simpleLoess(y, x, w, span, degree = degree, parametric =
># parametric, : reciprocal condition number 4.3494e-30># Warning in simpleLoess(y, x, w, span, degree = degree, parametric =
># parametric, : There are other near singularities as well. 1.01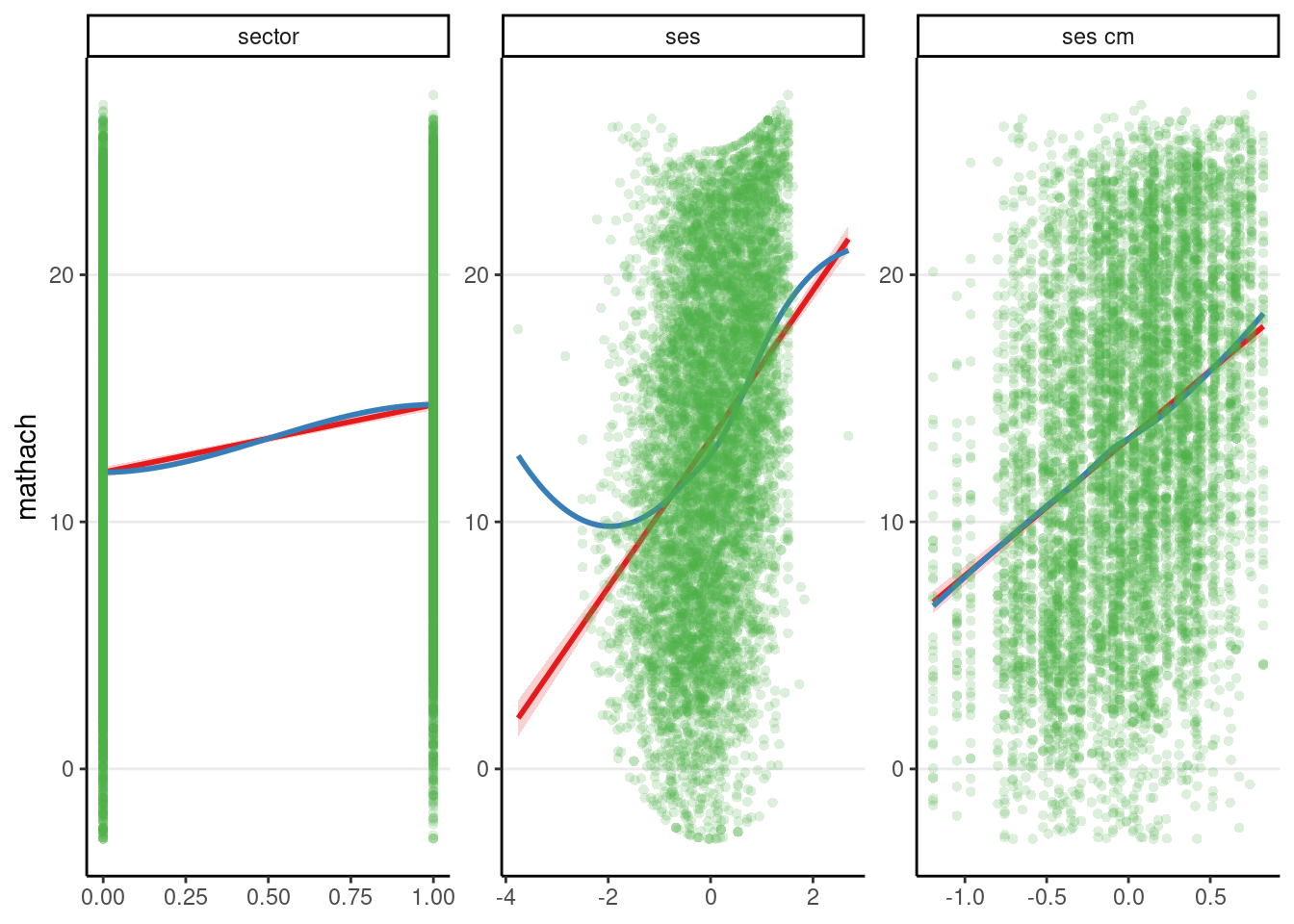
The above shows a misfit for the plot with ses.
What to do if linearity is violated?
- Include nonlinear terms (e.g., quadratic)
- Include interactions (but note that interactions can be present even when linearity is not violated)
- Using splines (see section 15.2.2 of Snijders & Bosker, 2012, or this paper for an introduction). Below is an example:
# Need the `spline` package; bs() for cubic spline
m2_spline <- lmer(mathach ~ sector + bs(ses) + bs(ses_cm) + (1 | id),
data = hsball
)
# See whether the nonlinear trend is needed
anova(m2, m2_spline) # not statistically significant># refitting model(s) with ML (instead of REML)npar <dbl> | AIC <dbl> | BIC <dbl> | logLik <dbl> | deviance <dbl> | Chisq <dbl> | ||
|---|---|---|---|---|---|---|---|
| m2 | 6 | 46560.32 | 46601.60 | -23274.16 | 46548.32 | NA | |
| m2_spline | 10 | 46565.01 | 46633.81 | -23272.50 | 46545.01 | 3.311813 |
# So insufficient evidence for a nonlinear trend for `ses` and `ses_cm`Does the random part contain the right variables?
Testing for random slopes
m3 <- lmer(mathach ~ sector + ses + ses_cm + (ses | id),
data = hsball)
ranova(m3) # remember to divide p by 2npar <int> | logLik <dbl> | AIC <dbl> | LRT <dbl> | Df <dbl> | ||
|---|---|---|---|---|---|---|
| <none> | 8 | -23273.74 | 46563.47 | NA | NA | |
| ses in (ses | id) | 6 | -23276.92 | 46565.83 | 6.361442 | 2 |
The random slope variance for ses was significant, so it should be included.
Residual Plots
Residuals can be used to check a lot of assumptions, including linearity, homoscedasticity, normality, and outliers.
Level-1 residuals across predictors
The plot below shows that heteroscedasticity is likely not a problem, as we do not see clearly bigger/smaller variances at certain levels of the predictors.
# Compute OLS residuals
m3_resid <- augment(m3) %>%
# The OLS residuals also capture the random effects (u's)
mutate(.ols_resid = mathach - .fixed)
# SES
ggplot(m3_resid, aes(x = ses, y = .ols_resid)) +
geom_point(size = 0.7, alpha = 0.5) +
geom_smooth(se = FALSE)># `geom_smooth()` using method = 'gam' and formula = 'y ~ s(x, bs = "cs")'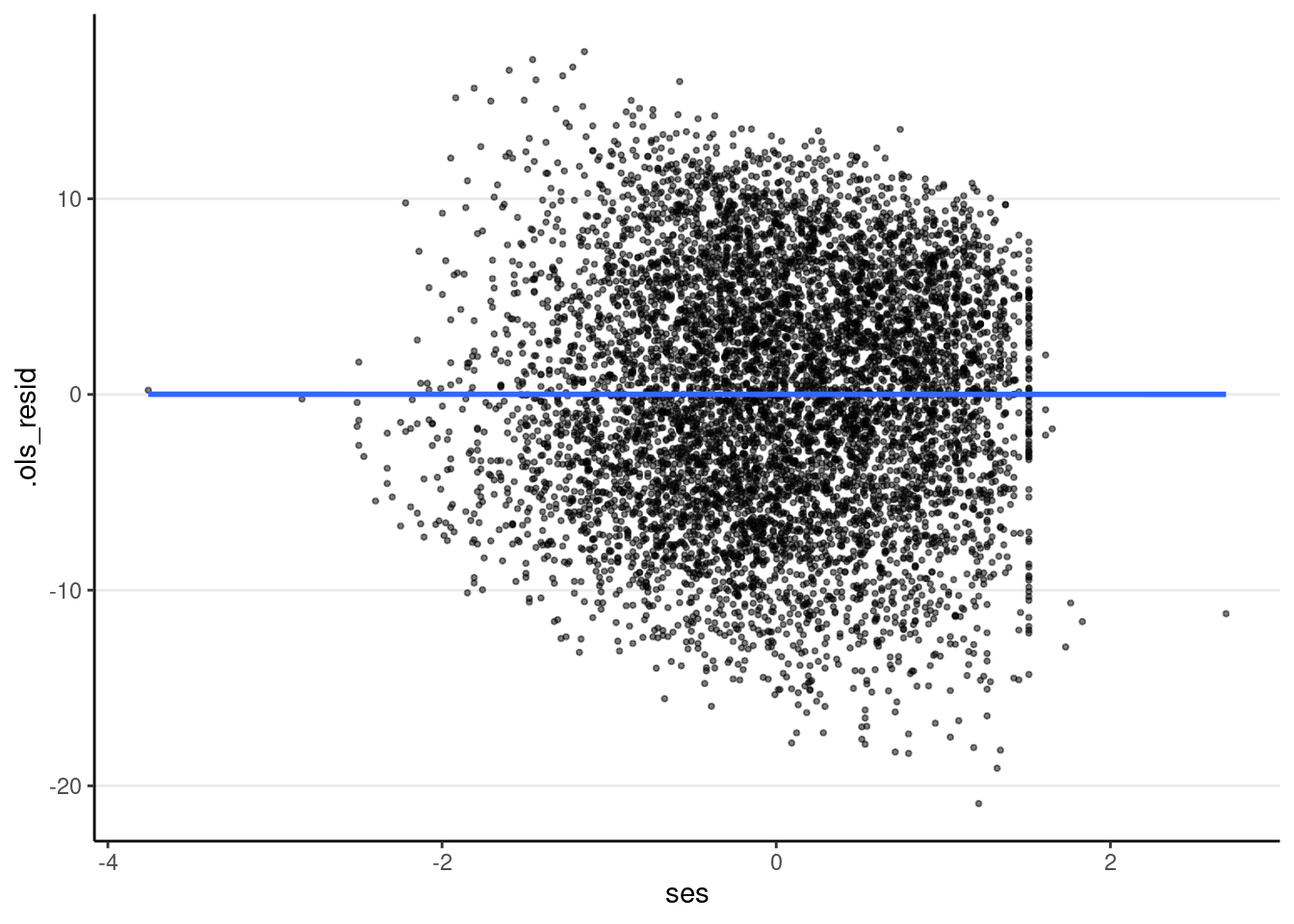
# ses_cm
ggplot(m3_resid, aes(x = ses_cm, y = .ols_resid)) +
geom_point(size = 0.7, alpha = 0.5) +
geom_smooth(se = FALSE)># `geom_smooth()` using method = 'gam' and formula = 'y ~ s(x, bs = "cs")'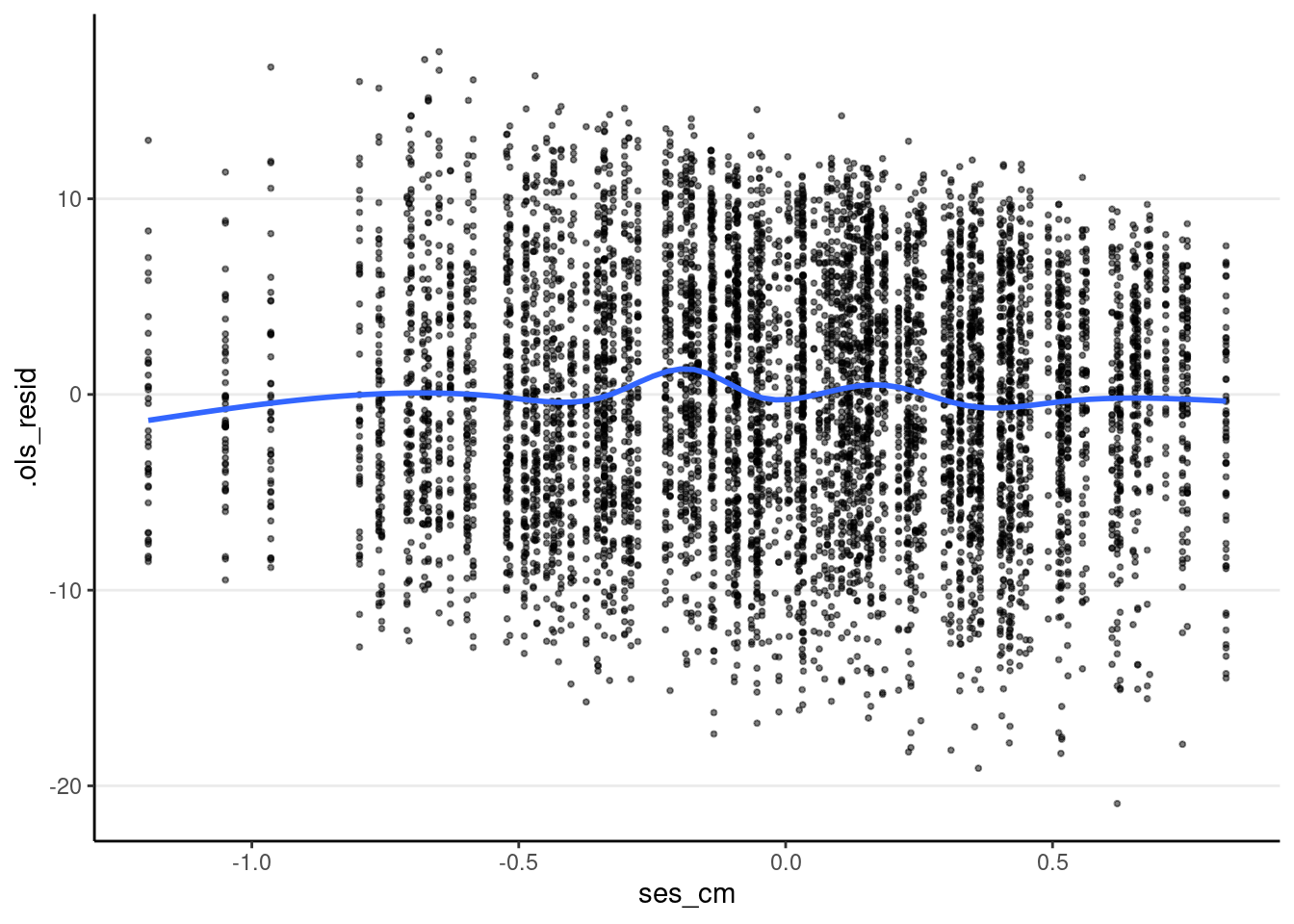
# sector
ggplot(m3_resid, aes(x = sector, y = .ols_resid)) +
# use `geom_jitter` for discrete predictor
geom_jitter(size = 0.7, alpha = 0.2, width = 0.05)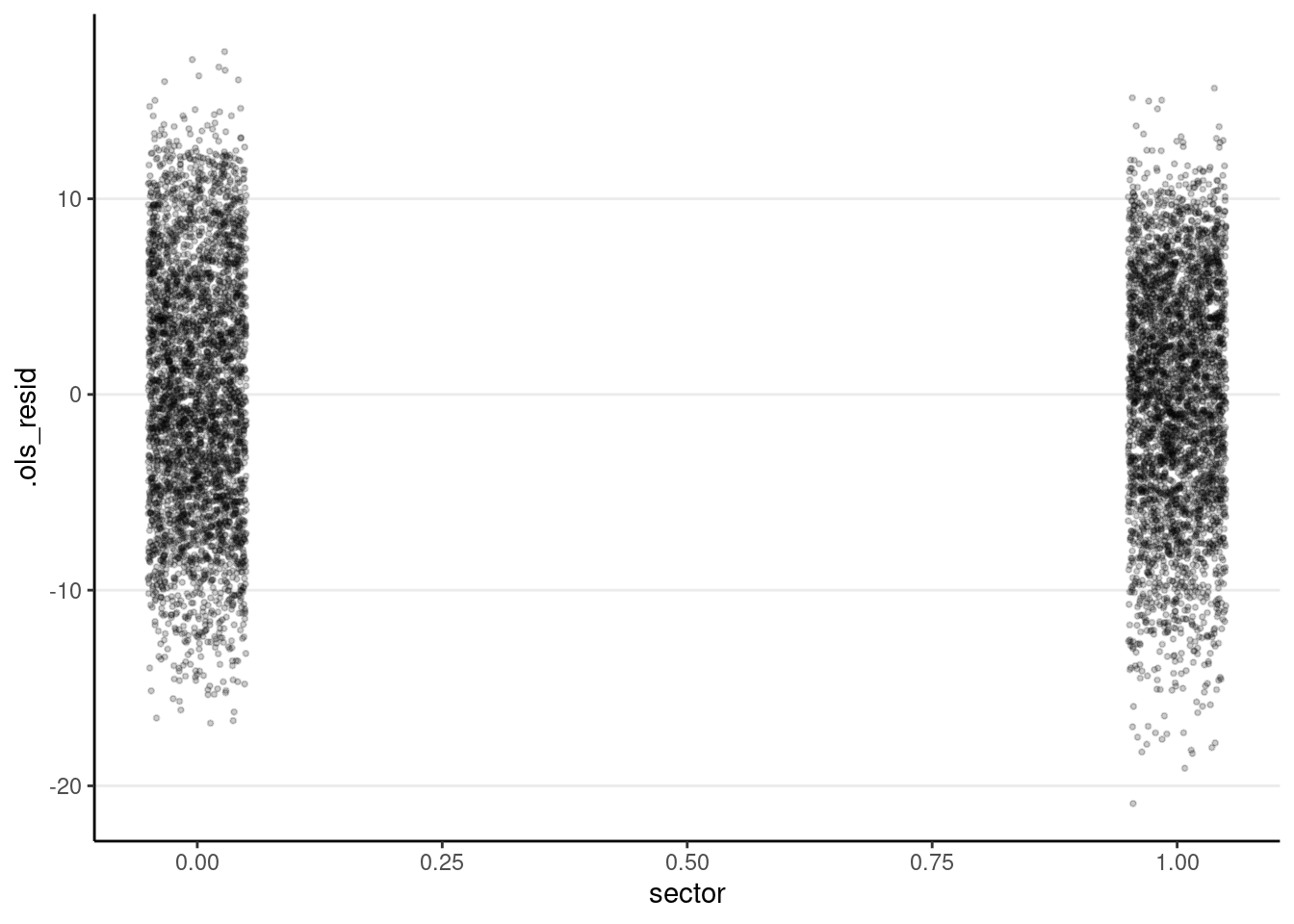
Level-2 residuals across lv-2 predictors
# Use `ranef()` to get lv-2 residuals
m3_resid_lv2 <- ranef(m3)$id # id is the name for the cluster variable
# Rename
names(m3_resid_lv2) <- c("u0j_hat", "u1j_hat")
# Data of lv-2 predictors (160 rows)
hsball_lv2 <- hsball %>%
group_by(id) %>%
summarize(ses_cm = ses_cm[1],
sector = sector[1]) %>%
ungroup()
# Combine residuals and predictors
m3_resid_lv2 <- cbind(m3_resid_lv2, hsball_lv2)
# Plot lv-2 residual against `ses_cm`
ggplot(m3_resid_lv2, aes(x = ses_cm, y = u0j_hat)) +
geom_point(size = 0.7, alpha = 0.5) +
geom_smooth(se = FALSE)># `geom_smooth()` using method = 'loess' and formula = 'y ~ x'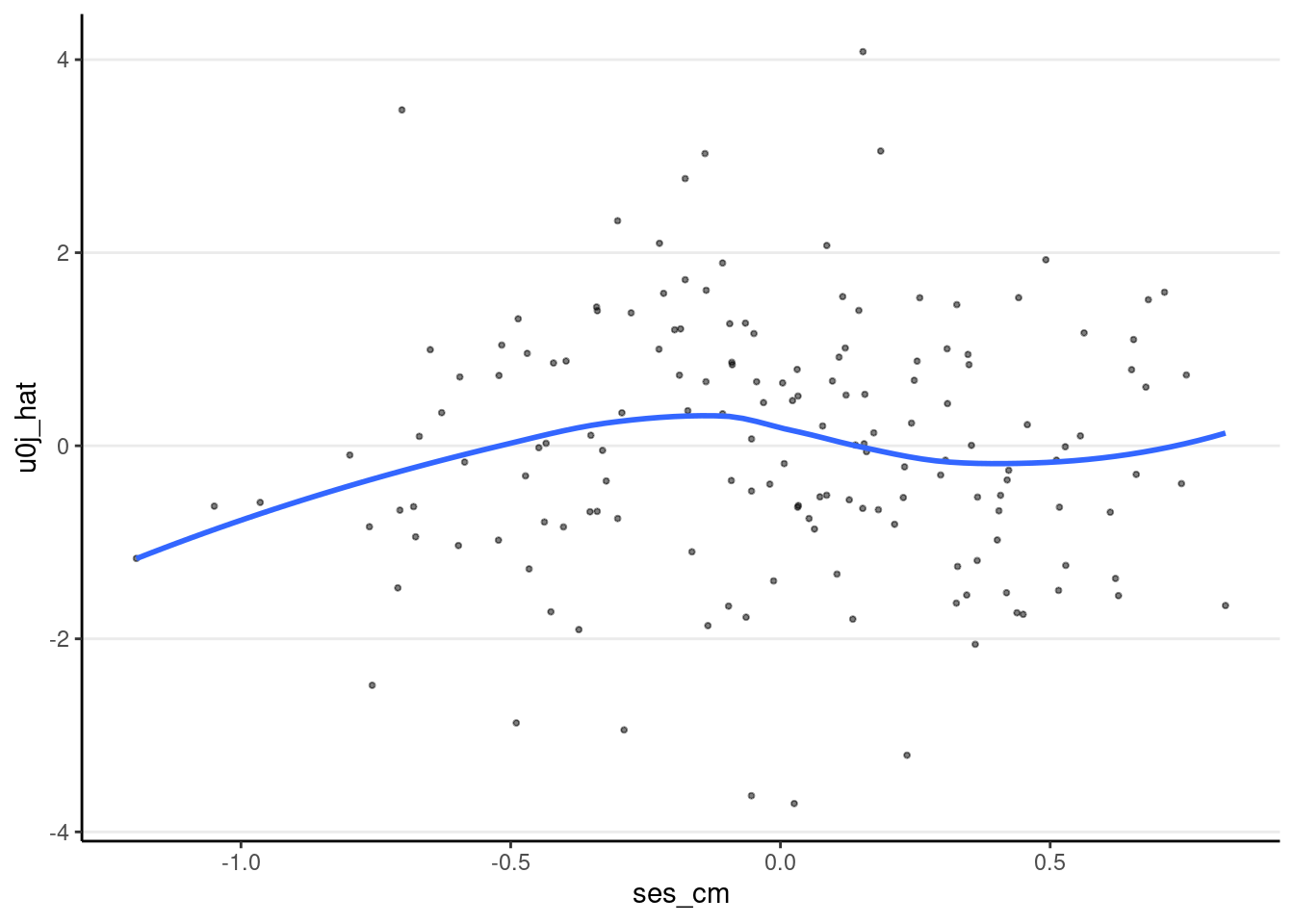
ggplot(m3_resid_lv2, aes(x = ses_cm, y = u1j_hat)) +
geom_point(size = 0.7, alpha = 0.5) +
geom_smooth(se = FALSE)># `geom_smooth()` using method = 'loess' and formula = 'y ~ x'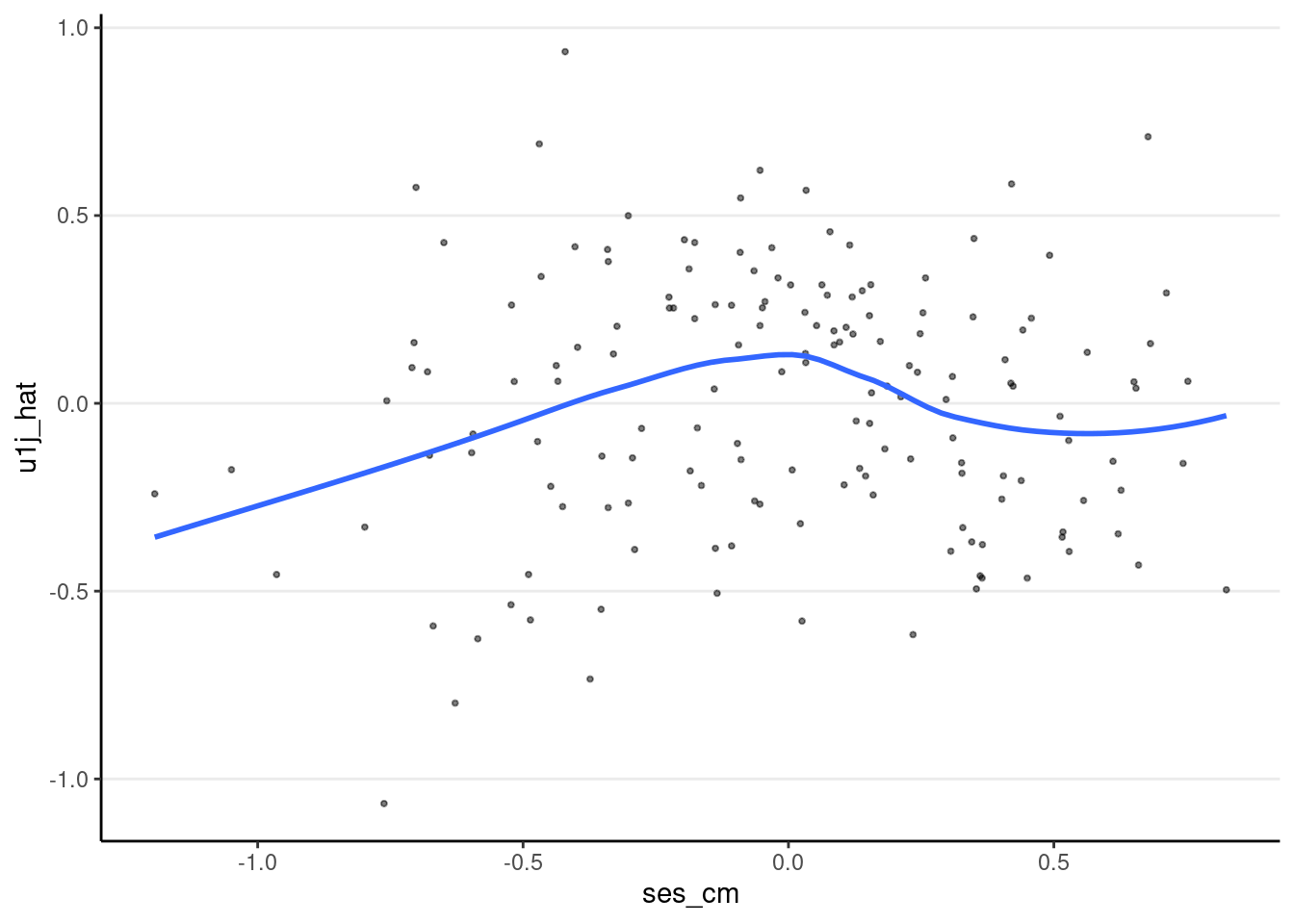
# Plot lv-2 residual against `sector`
ggplot(m3_resid_lv2, aes(x = sector, y = u0j_hat)) +
geom_jitter(size = 0.7, alpha = 0.5, width = 0.05)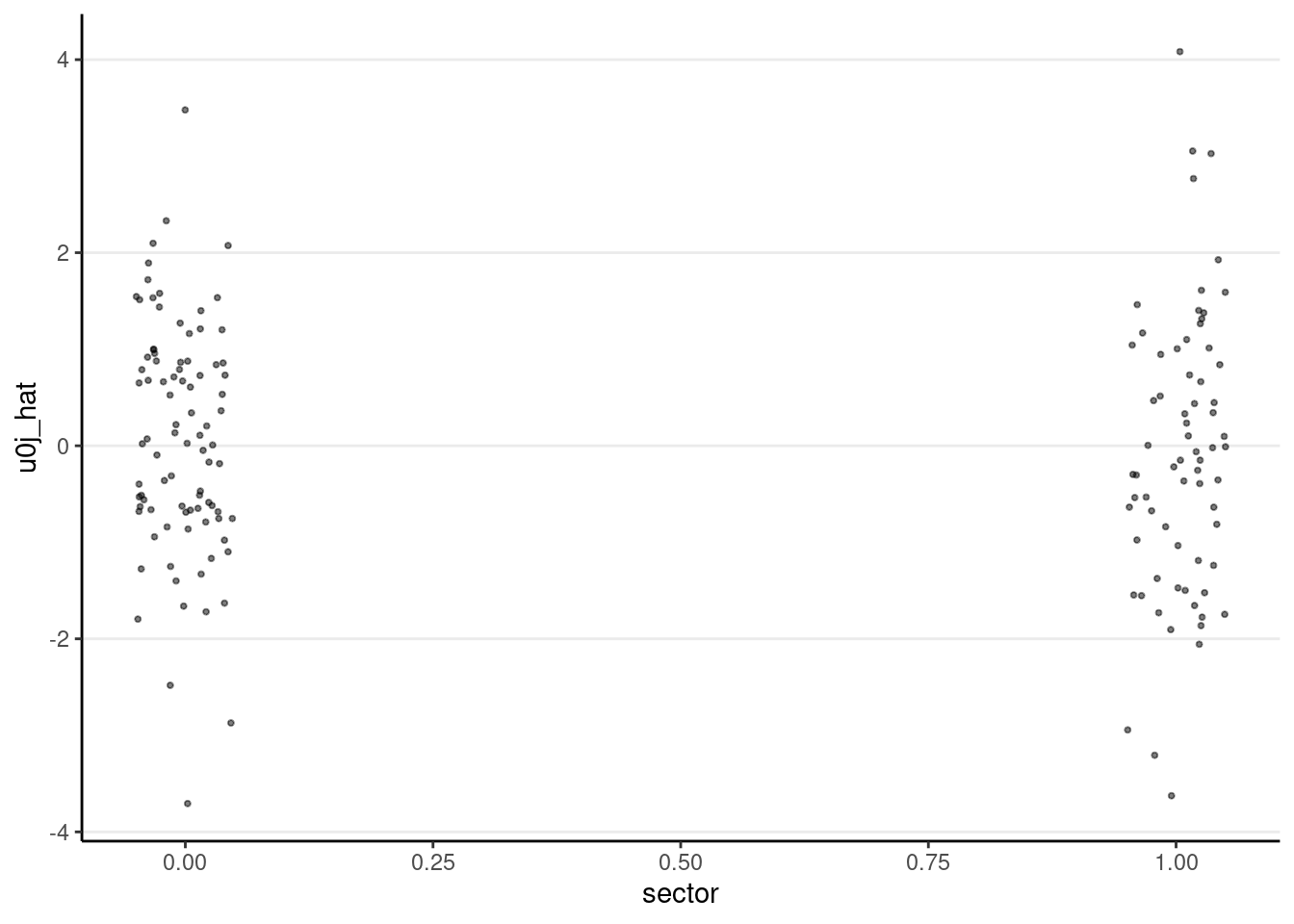
ggplot(m3_resid_lv2, aes(x = sector, y = u1j_hat)) +
geom_jitter(size = 0.7, alpha = 0.5, width = 0.05)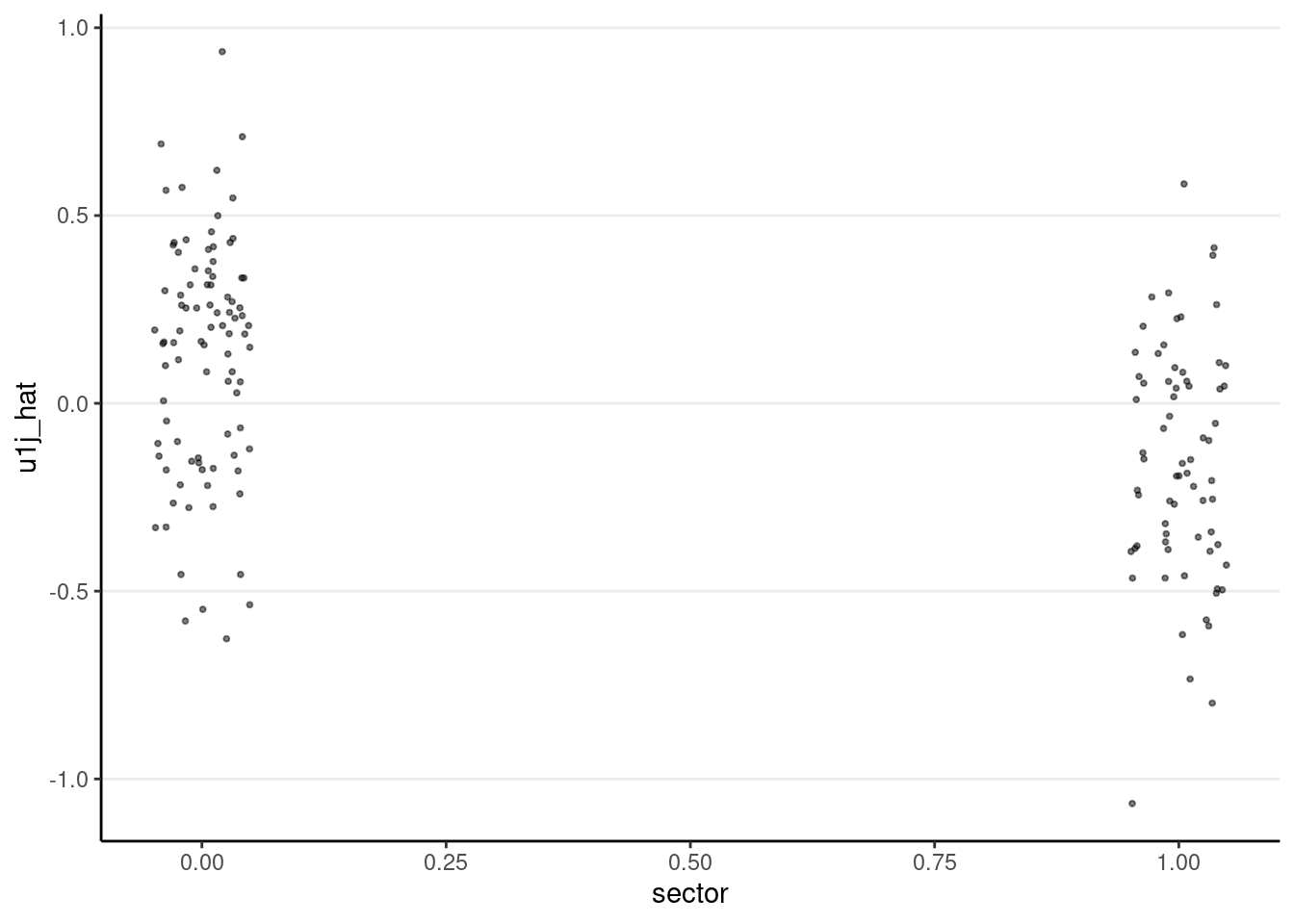
Bonus: Heteroscedasticity-consistent (HC) variance estimates
Suppose you suspect that heteroscedasticity may be a problem. In that case, you can also use the heteroskedasticity-consistent variance estimators with the clubSandwich package, which computes the HC correction for the test results.
library(clubSandwich)># Registered S3 method overwritten by 'clubSandwich':
># method from
># bread.mlm sandwich# Sampling variances of fixed effects from REML
vcov_m3 <- vcov(m3)
# Sampling variances from HC
vcovcr_m3 <- vcovCR(m3, type = "CR2")
# Compare standard errors: REML-SE and HC-SE
cbind(
"REML-SE" = sqrt(diag(vcov_m3)),
"HC-SE" = sqrt(diag(vcovcr_m3))
)># REML-SE HC-SE
># (Intercept) 0.2016592 0.1779908
># sector 0.3090955 0.3157639
># ses 0.1223825 0.1287679
># ses_cm 0.3880834 0.3659749# Testing fixed effects adjusting for heteroscedasticity
coef_test(m3, vcov = "CR2")Coef <chr> | beta <dbl> | SE <dbl> | tstat <dbl> | ||
|---|---|---|---|---|---|
| (Intercept) | (Intercept) | 12.043386 | 0.1779908 | 67.662977 | |
| sector | sector | 1.408363 | 0.3157639 | 4.460178 | |
| ses | ses | 2.196100 | 0.1287679 | 17.054712 | |
| ses_cm | ses_cm | 3.155202 | 0.3659749 | 8.621364 |
Note: To my knowledge, currently it is not possible to combine the HC estimator with the small-sample Kenward-Roger correction.
Q-Q Plot for Residuals
For: normality
Lv-1
You want to see the points line up nicely along the 45-degree line.
library(lattice) # need this package to use the built-in functions
qqmath(m3) # just use the `qqmath()` function on the fitted model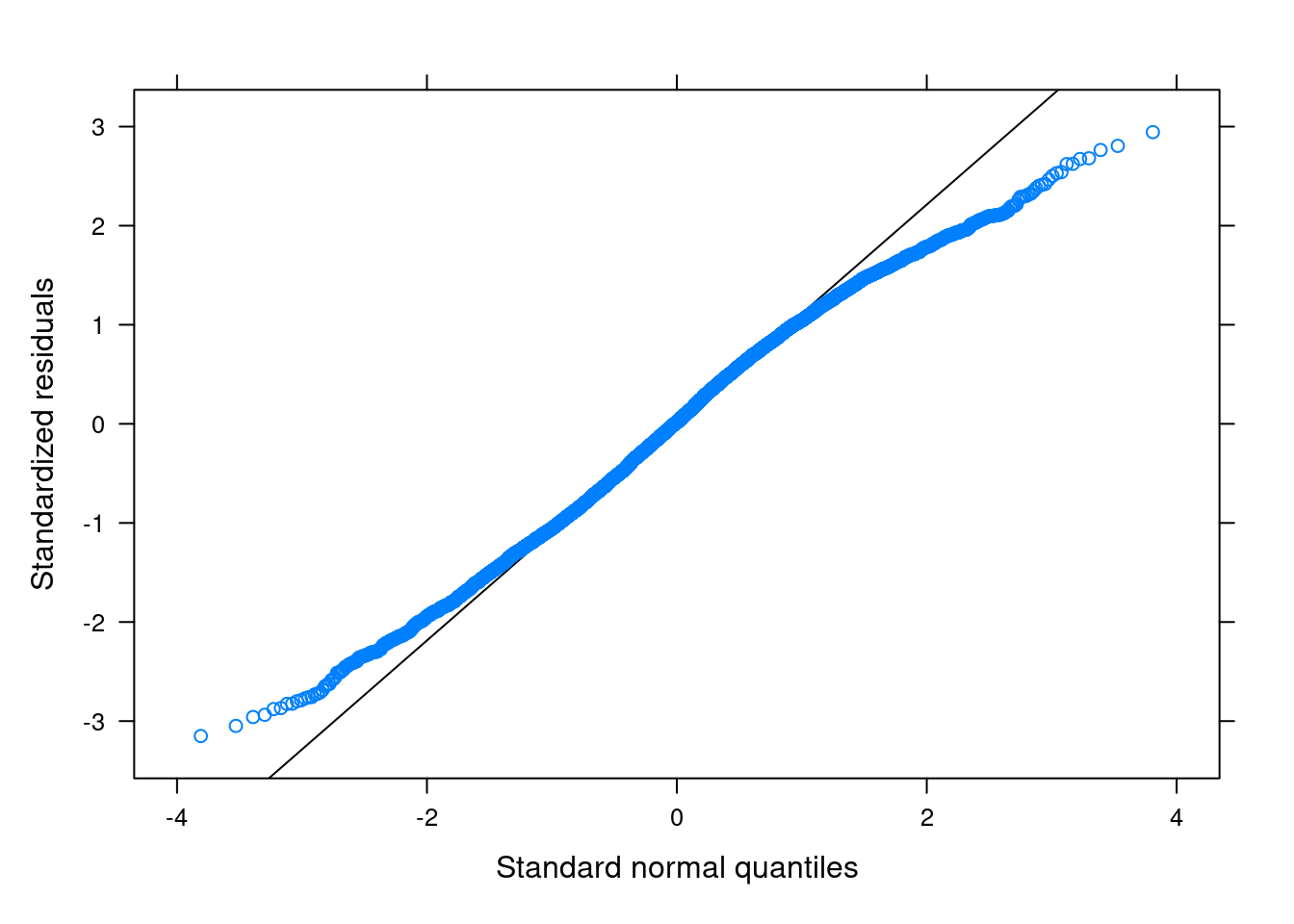
There are some deviations from normality in the tail areas, likely due to the boundedness of the outcome variable. Generally, the impact of kurtosis is not as severe as skewness, and the above plot did not suggest a major issue to the results.
Lv-2
At level 2, there can be more than one set of residuals. For example, in this model, we have one set for the random intercepts, and the other for the random slopes.
# First, get standardized residuals
resid_lv2 <- ranef(m3)
# The code below gets the standard error of the residuals
resid_lv2_var <- attr(resid_lv2$id, "postVar")
resid_lv2_sd <- sqrt(cbind(
resid_lv2_var[1, 1, ],
resid_lv2_var[2, 2, ]
))
# Standardized residuals = residuals / standard errors
std_resid_lv2 <- resid_lv2
std_resid_lv2$id <- std_resid_lv2$id / resid_lv2_sd
# Q-Q plot
qqmath(std_resid_lv2,
panel = function(x) {
panel.qqmath(x)
panel.qqmathline(x)
})># $id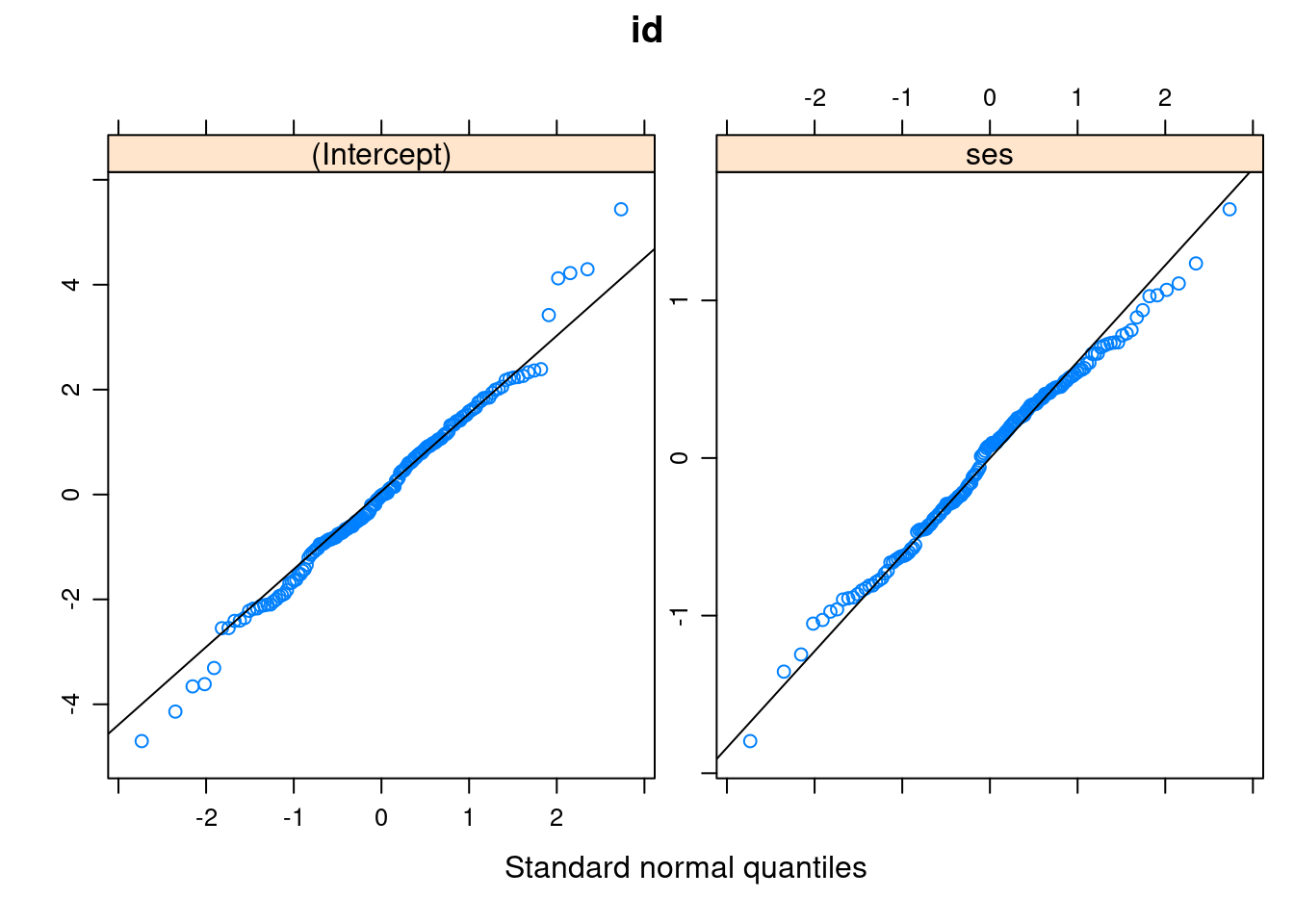
Normality does not appear to be an issue at level 2.
Bonus: Variance Inflation Factor (VIF)
For: multicollinearity
library(car)
car::vif(m3)># sector ses ses_cm
># 1.135375 1.081972 1.217836As a rule of thumb, one should worry about multicollinearity in the predictors when VIF > 4, and VIF > 10 indicates strong multicollinearity.
Bonus: Influence Index Plots
For: leverage points and outliers
To examine whether some cases have a disproportionately strong influence on the results, we can look at the influence index plot and dfbeta.
library(car)
inf_m3 <- influence(m3, "id") # get cluster-level influence statistics
car::infIndexPlot(inf_m3)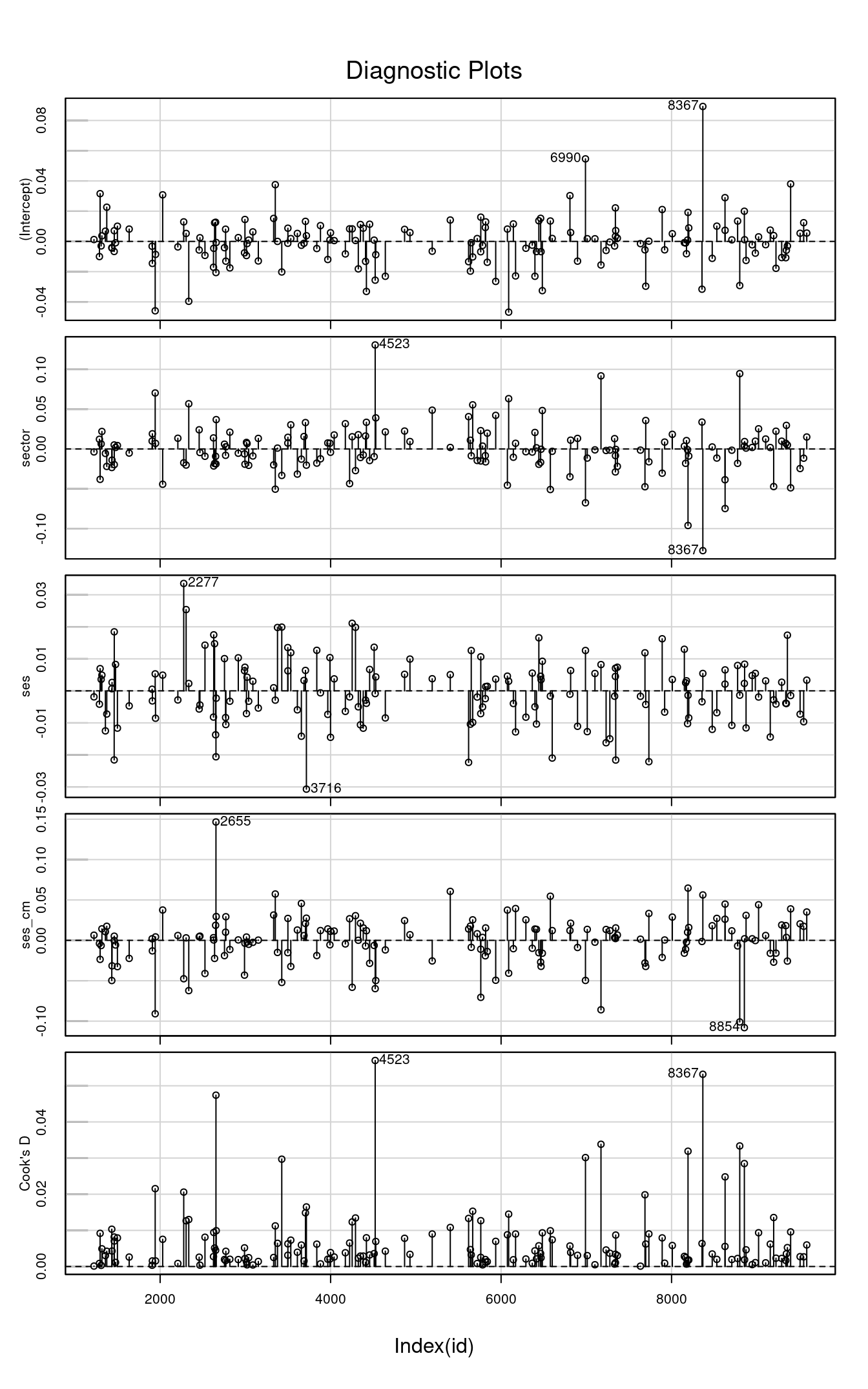
If you need some cutoffs, the cutoff for dfbeta is
Bonus: Bayesian Robust MLM
There has been a limited amount of discussion on MLM that is robust to outliers. One useful technique, however, is to use a heavy-tailed distribution for the errors and random effects. Instead of assuming that they follow normal distributions, we assume they follow Student’s brms package (see note in Week 5 for installing this package).
library(brms)># Loading required package: Rcpp># Loading 'brms' package (version 2.18.0). Useful instructions
># can be found by typing help('brms'). A more detailed introduction
># to the package is available through vignette('brms_overview').>#
># Attaching package: 'brms'># The following object is masked from 'package:lme4':
>#
># ngrps># The following object is masked from 'package:stats':
>#
># ar# Same syntax; just change `lmer()` to `brm()`
m3_robust <- brm(
mathach ~ sector + ses + ses_cm,
# Assume the errors follow a t distribution
family = student(),
data = hsball,
# You usually need to set `adapt_delta` to close to 1 (like .95 or .99)
# for robust models
control = list(adapt_delta = .95),
file = "brms_cached_m3_robust"
)The results are similar, so the regular MLM should be fine here.
msummary(
list(`Normal MLM` = m3,
`Robust MLM` = m3_robust),
estimate = "{estimate} [{conf.low}, {conf.high}]",
statistic = NULL,
metrics = "none"
)># Loading required namespace: rstanarm| Normal MLM | Robust MLM | |
|---|---|---|
| (Intercept) | 12.043 [11.648, 12.439] | |
| sector | 1.408 [0.802, 2.014] | |
| ses | 2.196 [1.956, 2.436] | |
| ses_cm | 3.155 [2.394, 3.916] | |
| b_Intercept | 12.120 [11.903, 12.325] | |
| b_sector | 1.287 [0.980, 1.598] | |
| b_ses | 2.210 [1.997, 2.421] | |
| b_ses_cm | 2.980 [2.547, 3.422] | |
| sigma | 6.229 [6.124, 6.339] | |
| SD (Intercept id) | 1.559 | |
| SD (ses id) | 0.688 | |
| Cor (Intercept~ses id) | 0.270 | |
| SD (Observations) | 6.065 | |
| Num.Obs. | 7185 | 7185 |
| AIC | 46563.5 | |
| BIC | 46618.5 | |
| Log.Lik. | −23273.735 | |
| REMLcrit | 46547.471 | |
| algorithm | sampling | |
| pss | 4000.000 |
Convergence
It’s not uncommon to run into convergence issues when the model contains two or more random slopes, or when the sample size (i.e., number of clusters) is small.
Multiple Random Slopes
Here I include female and its random slopes.
m_complex <- lmer(
mathach ~ sector + ses + ses_cm + female + (ses + female | id),
data = hsball
)># Warning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
># Model failed to converge with max|grad| = 0.00249344 (tol = 0.002, component 1)You will see the above warning. This almost always happens with two or more random slopes.
You can consult https://bbolker.github.io/mixedmodels-misc/glmmFAQ.html#troubleshooting regarding some suggestions on what to do (and I recommend that you save this page for your future reference).
The first thing you can do is to do centering and scaling. Scaling means making the predictors have a smaller range. For example, if one of your predictors is income in dollars, I suggest dividing income by $10,000 so that the resulting values are smaller. For centering, we can try using the cluster-mean centered variables:
# Cluster-mean centering
hsball <- hsball %>%
group_by(id) %>%
mutate(female_cm = mean(female),
female_cmc = female - female_cm) %>%
ungroup() # exit the "editing within groups" modem_complex2 <- lmer(
mathach ~ sector + ses_cmc + ses_cm + female_cmc + female_cm +
(ses_cmc + female_cmc | id),
data = hsball
)The warning actually goes away now. If it does not, the next thing you can do is to try a different optimizer:
m_complex_all <- lme4::allFit(m_complex)># Loading required namespace: dfoptim># Loading required namespace: optimx># bobyqa :># boundary (singular) fit: see help('isSingular')># Warning: Model failed to converge with 2 negative eigenvalues: -8.4e-04 -9.5e+03># [OK]
># Nelder_Mead : [OK]
># nlminbwrap : [OK]
># nmkbw : [OK]
># optimx.L-BFGS-B :># boundary (singular) fit: see help('isSingular')># Warning: Model failed to converge with 2 negative eigenvalues: -5.0e-01 -7.0e+03># [OK]
># nloptwrap.NLOPT_LN_NELDERMEAD : [OK]
># nloptwrap.NLOPT_LN_BOBYQA :># Warning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
># Model failed to converge with max|grad| = 0.00249344 (tol = 0.002, component 1)># [OK]ss <- summary(m_complex_all) # this shows everything
# Check which optimizer work
ss$which.OK># bobyqa Nelder_Mead
># TRUE TRUE
># nlminbwrap nmkbw
># TRUE TRUE
># optimx.L-BFGS-B nloptwrap.NLOPT_LN_NELDERMEAD
># TRUE TRUE
># nloptwrap.NLOPT_LN_BOBYQA
># TRUE# Compare the converged log-likelihood from different optimizers
ss$llik># bobyqa Nelder_Mead
># -23314.34 -23244.43
># nlminbwrap nmkbw
># -23244.43 -23244.43
># optimx.L-BFGS-B nloptwrap.NLOPT_LN_NELDERMEAD
># -23314.34 -23244.43
># nloptwrap.NLOPT_LN_BOBYQA
># -23244.43# Compare the fixed effects
ss$fixef># (Intercept) sector ses ses_cm female
># bobyqa 12.81487 1.279043 2.160235 3.104912 -1.344823
># Nelder_Mead 12.66714 1.376844 2.154596 3.075339 -1.192094
># nlminbwrap 12.66709 1.376950 2.154601 3.075310 -1.192104
># nmkbw 12.66716 1.376797 2.154596 3.075361 -1.192100
># optimx.L-BFGS-B 12.81496 1.278858 2.160219 3.104946 -1.344837
># nloptwrap.NLOPT_LN_NELDERMEAD 12.66713 1.376858 2.154597 3.075339 -1.192099
># nloptwrap.NLOPT_LN_BOBYQA 12.66716 1.376774 2.154590 3.075422 -1.192069So it looks like the bobyqa optimizer (the default) converges to a different solution than other optimizers. The log-likelihood value is better (i.e., less negative) for the other optimizers than the default, so we should try those. Let’s use nlminbwrap
m_complex <- lmer(
mathach ~ sector + ses + ses_cm + female + (ses + female | id),
data = hsball,
control = lmerControl(optimizer = "nlminbwrap")
)The warning goes away.
Singular Models
Especially in small samples, you will run into a message of singular model fit, like
# Randomly select 10 school ids
set.seed(1059) # use the same seed so that the same 10 schools are selected
random_ids <- sample(unique(hsball$id), size = 10)
hsbsub <- hsball %>%
filter(id %in% random_ids)
# Fit a model
m3_small <- lmer(mathach ~ sector + ses + ses_cm + (ses | id),
data = hsbsub)># boundary (singular) fit: see help('isSingular')summary(m3_small)># Linear mixed model fit by REML. t-tests use Satterthwaite's method [
># lmerModLmerTest]
># Formula: mathach ~ sector + ses + ses_cm + (ses | id)
># Data: hsbsub
>#
># REML criterion at convergence: 2798.7
>#
># Scaled residuals:
># Min 1Q Median 3Q Max
># -2.56377 -0.69920 -0.03495 0.74830 2.88703
>#
># Random effects:
># Groups Name Variance Std.Dev. Corr
># id (Intercept) 3.0198 1.7377
># ses 0.2445 0.4944 -1.00
># Residual 40.1346 6.3352
># Number of obs: 428, groups: id, 10
>#
># Fixed effects:
># Estimate Std. Error df t value Pr(>|t|)
># (Intercept) 11.944 0.875 6.904 13.650 3e-06 ***
># sector -1.018 1.478 6.342 -0.689 0.515508
># ses 2.247 0.473 18.902 4.750 0.000141 ***
># ses_cm 3.701 2.322 8.475 1.594 0.147534
># ---
># Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
>#
># Correlation of Fixed Effects:
># (Intr) sector ses
># sector -0.663
># ses -0.251 0.045
># ses_cm 0.480 -0.478 -0.240
># optimizer (nloptwrap) convergence code: 0 (OK)
># boundary (singular) fit: see help('isSingular')If you look at the random effect matrix, the correlation between the random intercepts and the random slopes is -1, which is small. Usually, this is not a big problem for the fixed effects. If you have a highly complex model with a small sample, then the recommendation is to simplify the model by taking out one of the random slope terms. An alternative is to use Bayesian estimation, which is less prone to small-sample estimation problems. For example, I will use the brms package.
# Same syntax; just replace `lmer` with `brm`
m3_small_bayes <- brm(
mathach ~ sector + ses + ses_cm + (ses | id),
data = hsbsub,
file = "brms_cached_m3_small_bayes"
)msummary(
list(REML = m3_small,
Bayes = m3_small_bayes),
estimate = "{estimate} [{conf.low}, {conf.high}]",
statistic = NULL,
metrics = "none"
)| REML | Bayes | |
|---|---|---|
| (Intercept) | 11.944 [10.224, 13.664] | |
| sector | −1.018 [−3.924, 1.888] | |
| ses | 2.247 [1.317, 3.177] | |
| ses_cm | 3.701 [−0.863, 8.266] | |
| b_Intercept | 11.607 [9.284, 13.909] | |
| b_sector | −0.352 [−4.351, 3.793] | |
| b_ses | 2.209 [1.103, 3.233] | |
| b_ses_cm | 3.000 [−3.060, 8.742] | |
| sd_id__Intercept | 2.042 [0.918, 4.313] | |
| sd_id__ses | 0.632 [0.029, 2.211] | |
| cor_id__Intercept__ses | −0.244 [−0.961, 0.904] | |
| sigma | 6.353 [5.941, 6.806] | |
| SD (Intercept id) | 1.738 | |
| SD (ses id) | 0.494 | |
| Cor (Intercept~ses id) | −1.000 | |
| SD (Observations) | 6.335 | |
| Num.Obs. | 428 | 428 |
| AIC | 2814.7 | |
| BIC | 2847.1 | |
| Log.Lik. | −1399.335 | |
| REMLcrit | 2798.669 | |
| algorithm | sampling | |
| pss | 4000.000 |
If the two are similar, then the REML results should be fine. Otherwise, as is the case here, the Bayesian result should be more trustworthy.