# To install a package, run the following ONCE (and only once on your computer)
# install.packages("psych")
library(here) # makes reading data more consistent
library(tidyverse) # for data manipulation and plotting
library(haven) # for importing SPSS/SAS/Stata data
library(dagitty) # for DAG
library(ggdag) # for DAG in ggplot style
library(lme4) # for multilevel analysis
library(sjPlot) # for plotting
library(mediation) # for plotting
library(modelsummary) # for making tablesWeek 11 (R): Causal Inference
Load Packages and Import Data
Causal Diagram
You can use the daggity package to draw causal diagrams, specifically directed acyclic graphs (DAGs). The ggdag package further makes the DAG in ggplot style. The following shows a confounder C.
dag1 <- dagitty(
"dag{
X -> Y; C -> X; C -> Y
}"
)
coordinates(dag1) <- list(x = c(X = 0, C = 1, Y = 2),
y = c(X = 0, C = 1, Y = 0))
# Plot
ggdag(dag1) + theme_dag()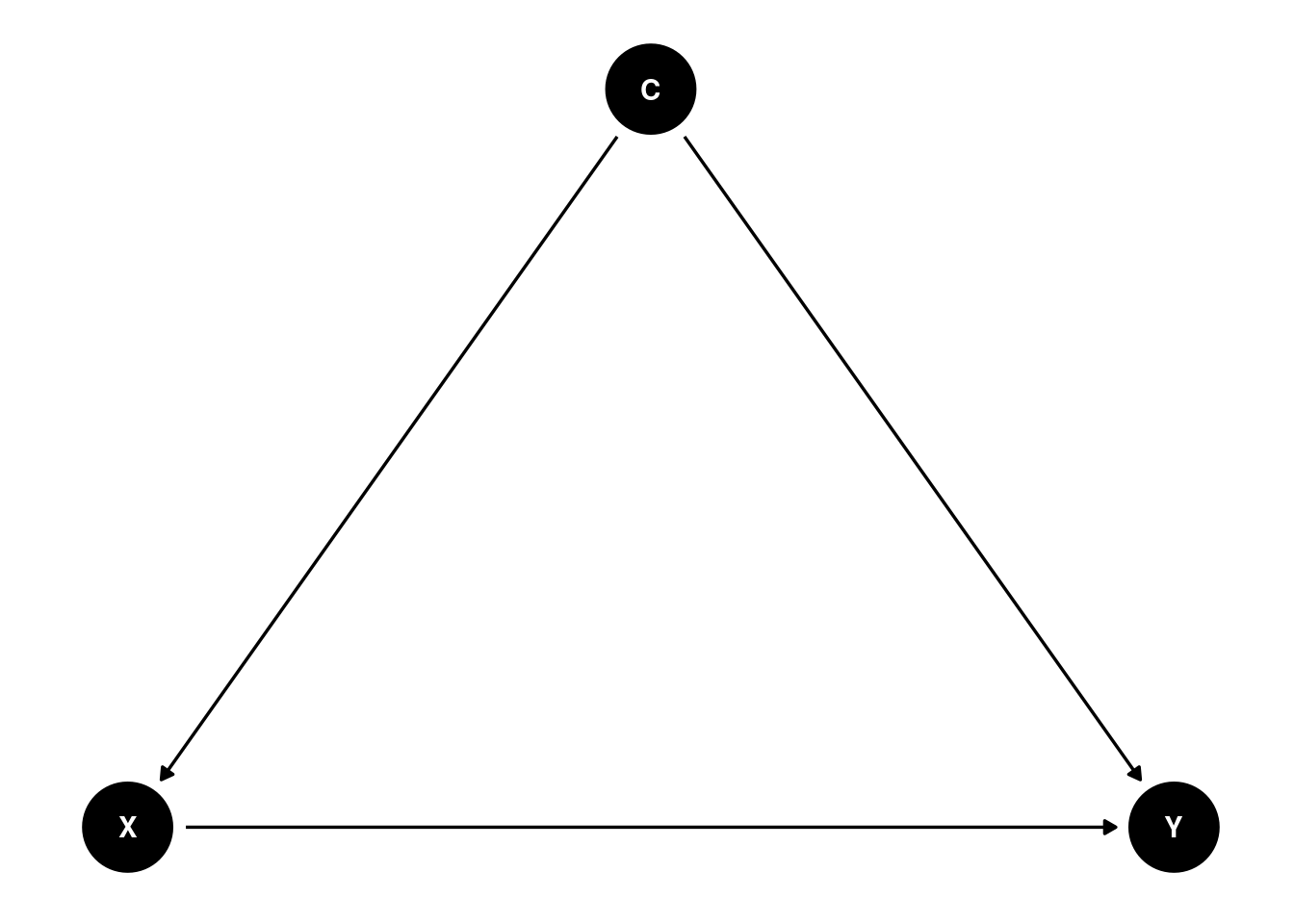
The dagitty package is very powerful, as it can inform what covariates need to be adjusted for a given causal diagram. This requires strong assumptions on the causal structure of the data. For example,
adjustmentSets(dag1, exposure = "X", outcome = "Y",
effect = "direct")># { C }tells you that C needs to be adjusted to obtain a causal effect of X to Y.
Statistical Adjustment
Import Data
# Import HSB data
hsball <- read_sav(here("data_files", "hsball.sav"))
# Cluster means
hsball <- hsball %>%
group_by(id) %>% # operate within schools
mutate(ses_cm = mean(ses),
minority_cm = mean(minority)) %>%
ungroup()Unadjusted and Adjusted Models
The observed difference between the two types of schools is not the causal effect of sector, namely,
What would the students perform if they changed from a public school to a Catholic school
when students can “select” to attend different schools. However, if we can obtain a complete set of variables that unconfounds the X → Y path, the adjusted coefficient can be interpreted as causal effects.
Assumptions:
- The covariates used completely unconfound all confounding paths
- The covariates used do not contain mediators or colliders
- All standard assumptions in regression and multilevel models (e.g., linearity)
Let’s say we think that the path is confounded because students with certain SES backgrounds may be more likely to go into public schools than Catholic schools. Also, minority students may be more likely to go to public schools. Both ses and minority are also on a causal path to mathach, and so both are on confounding paths of sector → mathach.
# Unadjusted model
m_unadjusted <- lmer(mathach ~ sector + (1 | id), data = hsball)
# Adjusted model (with ses and minority)
m_adjusted <- lmer(mathach ~ sector + ses_cm + minority_cm +
ses + minority + (ses + minority | id),
data = hsball)
msummary(list(unadjusted = m_unadjusted,
adjusted = m_adjusted))| unadjusted | adjusted | |
|---|---|---|
| (Intercept) | 11.393 | 12.682 |
| (0.293) | (0.213) | |
| sector | 2.805 | 1.545 |
| (0.439) | (0.303) | |
| ses_cm | 2.521 | |
| (0.431) | ||
| minority_cm | 0.096 | |
| (0.598) | ||
| ses | 1.936 | |
| (0.118) | ||
| minority | −2.945 | |
| (0.251) | ||
| SD (Intercept id) | 2.584 | 1.372 |
| SD (ses id) | 0.554 | |
| SD (minority id) | 1.195 | |
| Cor (Intercept~ses id) | 0.198 | |
| Cor (Intercept~minority id) | −0.053 | |
| Cor (ses~minority id) | −0.989 | |
| SD (Observations) | 6.257 | 5.981 |
| Num.Obs. | 7185 | 7185 |
| R2 Marg. | 0.041 | 0.209 |
| R2 Cond. | 0.181 | 0.261 |
| AIC | 47088.1 | 46375.3 |
| BIC | 47115.7 | 46464.7 |
| ICC | 0.1 | 0.07 |
| RMSE | 6.20 | 5.92 |
The estimated coefficient is smaller for the adjusted model. Note that this is only an example; it is unlikely that these two variables are sufficient to unconfound the associations between sector and mathach. For example, non-public schools may admit only students who already have a good math background, so we would need to adjust for a measure of math ability or aptitude.
Berkeley Admission Example
It is a famous data set, which is available in base R by calling
UCBAdmissions># , , Dept = A
>#
># Gender
># Admit Male Female
># Admitted 512 89
># Rejected 313 19
>#
># , , Dept = B
>#
># Gender
># Admit Male Female
># Admitted 353 17
># Rejected 207 8
>#
># , , Dept = C
>#
># Gender
># Admit Male Female
># Admitted 120 202
># Rejected 205 391
>#
># , , Dept = D
>#
># Gender
># Admit Male Female
># Admitted 138 131
># Rejected 279 244
>#
># , , Dept = E
>#
># Gender
># Admit Male Female
># Admitted 53 94
># Rejected 138 299
>#
># , , Dept = F
>#
># Gender
># Admit Male Female
># Admitted 22 24
># Rejected 351 317# ?UCBAdmissions # see documentationThe data, however, is a three-dimension contingency table, which cannot be directly analyzed by regression/multilevel models. Here we convert the data to a data frame:
berkeley_admit <- UCBAdmissions %>%
# to data frame
as.data.frame() %>%
# one column for admit and one for reject
pivot_wider(names_from = Admit, values_from = Freq)
berkeley_admitGender <fct> | Dept <fct> | Admitted <dbl> | Rejected <dbl> | |
|---|---|---|---|---|
| Male | A | 512 | 313 | |
| Female | A | 89 | 19 | |
| Male | B | 353 | 207 | |
| Female | B | 17 | 8 | |
| Male | C | 120 | 205 | |
| Female | C | 202 | 391 | |
| Male | D | 138 | 279 | |
| Female | D | 131 | 244 | |
| Male | E | 53 | 138 | |
| Female | E | 94 | 299 |
The outcome is the number of people admitted, which can be modelled as a binomial variable (i.e., number of “successes” out of a known total). We will talk about generalized linear models and generalized linear mixed models in a future week, but here’s the syntax when we ignore the department and just look at the overall success rate across Gender:
# For binomial, the outcome has the syntax `cbind(# successes, # failures)`
m_single <- glm(
cbind(Admitted, Rejected) ~ Gender,
data = berkeley_admit,
# specify family
family = binomial("logit")
)
summary(m_single)>#
># Call:
># glm(formula = cbind(Admitted, Rejected) ~ Gender, family = binomial("logit"),
># data = berkeley_admit)
>#
># Deviance Residuals:
># Min 1Q Median 3Q Max
># -16.7915 -4.7613 -0.4365 5.1025 11.2022
>#
># Coefficients:
># Estimate Std. Error z value Pr(>|z|)
># (Intercept) -0.22013 0.03879 -5.675 1.38e-08 ***
># GenderFemale -0.61035 0.06389 -9.553 < 2e-16 ***
># ---
># Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
>#
># (Dispersion parameter for binomial family taken to be 1)
>#
># Null deviance: 877.06 on 11 degrees of freedom
># Residual deviance: 783.61 on 10 degrees of freedom
># AIC: 856.55
>#
># Number of Fisher Scoring iterations: 4plot_model(m_single, type = "pred")># $Gender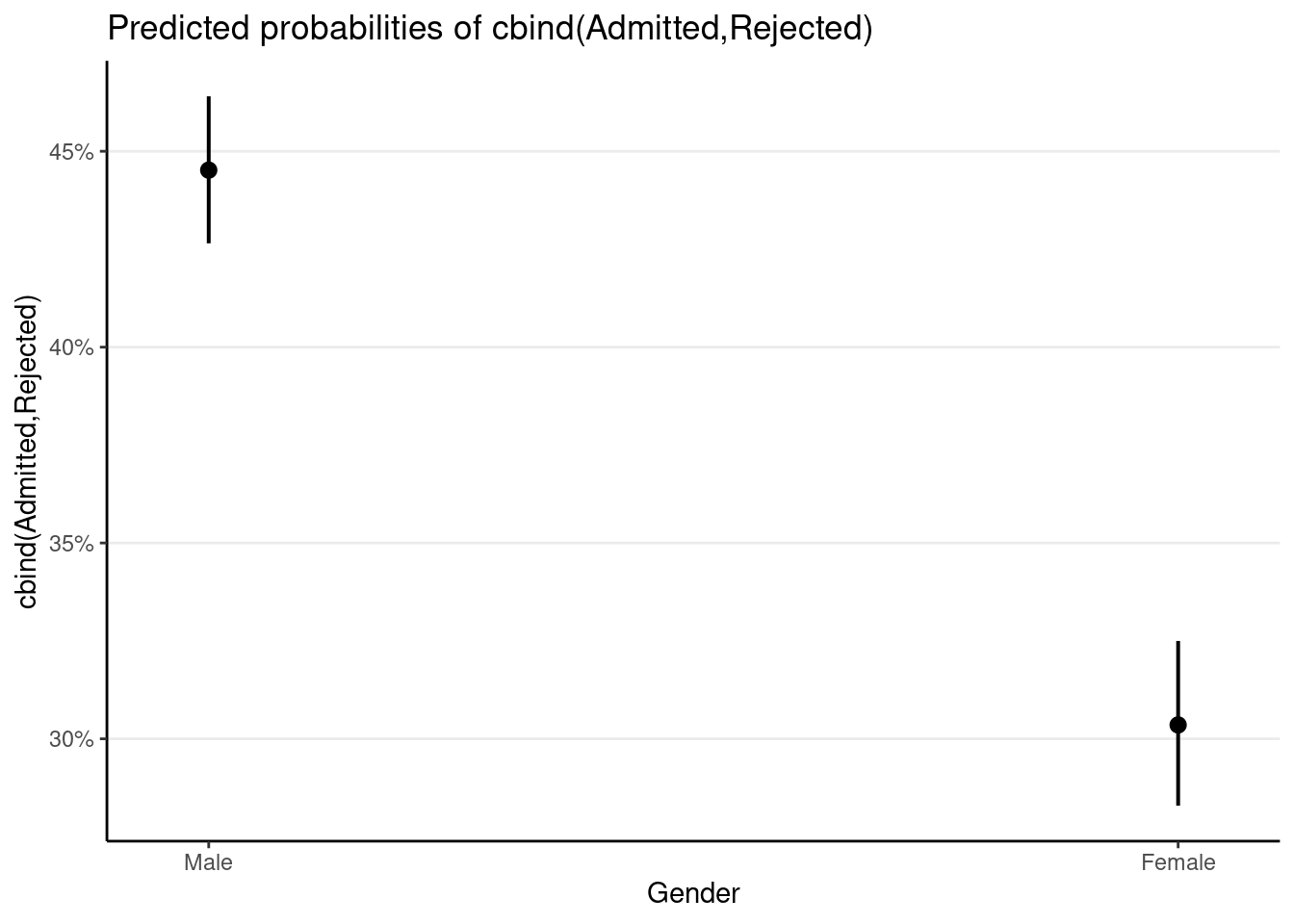
which shows a lower admission rate for females. However, if we also consider the department in a multilevel model, which also requires accounting for the proportion of female applicants in a given department, we have
# Cluster mean for Gender
berkeley_admit <- berkeley_admit |>
group_by(Dept) |>
mutate(
# Proportion of female applicants
Gender_cm = sum(Admitted[2], Rejected[2]) /
sum(Admitted, Rejected)) |>
ungroup()
berkeley_admitGender <fct> | Dept <fct> | Admitted <dbl> | Rejected <dbl> | Gender_cm <dbl> |
|---|---|---|---|---|
| Male | A | 512 | 313 | 0.11575563 |
| Female | A | 89 | 19 | 0.11575563 |
| Male | B | 353 | 207 | 0.04273504 |
| Female | B | 17 | 8 | 0.04273504 |
| Male | C | 120 | 205 | 0.64596950 |
| Female | C | 202 | 391 | 0.64596950 |
| Male | D | 138 | 279 | 0.47348485 |
| Female | D | 131 | 244 | 0.47348485 |
| Male | E | 53 | 138 | 0.67294521 |
| Female | E | 94 | 299 | 0.67294521 |
# Use `glmer()` for non-normal outcomes
m_mlm <- glmer(
cbind(Admitted, Rejected) ~ Gender + Gender_cm + (Gender | Dept),
data = berkeley_admit,
# specify family
family = binomial("logit")
)
summary(m_mlm)># Generalized linear mixed model fit by maximum likelihood (Laplace
># Approximation) [glmerMod]
># Family: binomial ( logit )
># Formula: cbind(Admitted, Rejected) ~ Gender + Gender_cm + (Gender | Dept)
># Data: berkeley_admit
>#
># AIC BIC logLik deviance df.resid
># 127.7 130.7 -57.9 115.7 6
>#
># Scaled residuals:
># Min 1Q Median 3Q Max
># -0.32605 -0.21671 -0.03468 0.14024 1.19665
>#
># Random effects:
># Groups Name Variance Std.Dev. Corr
># Dept (Intercept) 0.7428 0.8619
># GenderFemale 0.1131 0.3363 -0.14
># Number of obs: 12, groups: Dept, 6
>#
># Fixed effects:
># Estimate Std. Error z value Pr(>|z|)
># (Intercept) 0.6132 1.0576 0.580 0.562
># GenderFemale 0.1685 0.1722 0.978 0.328
># Gender_cm -3.1551 2.5040 -1.260 0.208
>#
># Correlation of Fixed Effects:
># (Intr) GndrFm
># GenderFemal 0.115
># Gender_cm -0.942 -0.181plot_model(m_mlm, terms = c("Dept [A, B, C, D, E, F]", "Gender"),
type = "pred", pred.type = "re")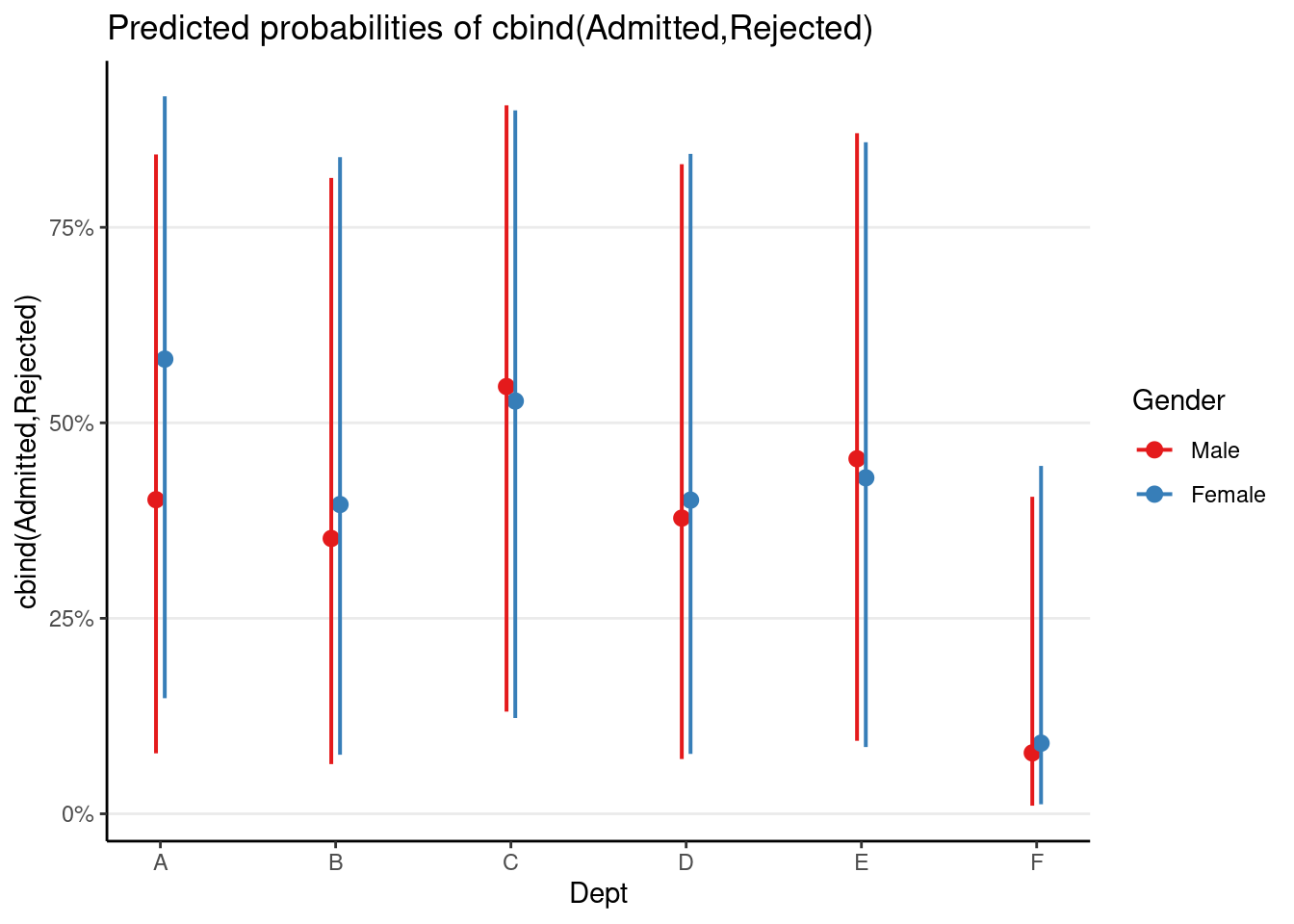
As can be seen, there was insufficient evidence for gender bias at the within-department level.
Bonus: Mediation
Here’s one of the examples from this paper (https://doi.org/10.3389/fpsyg.2020.00637), which reanalyzed some previous data on how beliefs influence judgments of learning (JOL) from Experiment 1 of Schaper et al. (2019). The mediation model here is Expectancy → ST (self-paced study time in seconds) → JOL. See this figure from the paper: https://www.frontiersin.org/files/Articles/522445/fpsyg-11-00637-HTML/image_m/fpsyg-11-00637-g002.jpg (the upper part).
Generally speaking, a mediation model has two components: (a) treatment to mediator, and (b) mediator to outcome, while holding the treatment constant. We can fit separate models to each component.
Import Data
dat <- haven::read_sav("https://osf.io/kewdc/download")
head(dat)sub <dbl> | expectancy <dbl> | diffbelief <dbl> | jol <dbl> | st_original <dbl> | st_log <dbl> | |
|---|---|---|---|---|---|---|
| 1 | -1 | -10 | 70 | 9641 | 9.173780 | |
| 1 | -1 | -10 | 80 | 3894 | 8.267192 | |
| 1 | -1 | -10 | 100 | 15223 | 9.630563 | |
| 1 | -1 | -10 | 70 | 6989 | 8.852093 | |
| 1 | -1 | -10 | 90 | 15950 | 9.677214 | |
| 1 | -1 | -10 | 70 | 14752 | 9.599134 |
dat$jol10 <- dat$jol / 10 # rescale JOL from 0-100 to 0-10
dat$belief10 <- dat$belief / 10 # rescale belief from 0-100 to 0-10Treatment → Mediator
Here the treatment is at level 1, and effect coding was used (-1 for control, 1 for treatment). With effect coding, the treatment coefficient is interpreted as the difference between a condition and the average, because one unit difference in the treatment variable means going from -1 to 0, where 0 is the average of the two conditions.
# Component (a). Cannot include random slope term in `lme4`
# with two observations per cluster. Can try glmmTMB(..., disp = ~0)
ma <- lmer(belief10 ~ expectancy + (1 | sub), data = dat)
plot_model(ma, type = "pred")># $expectancy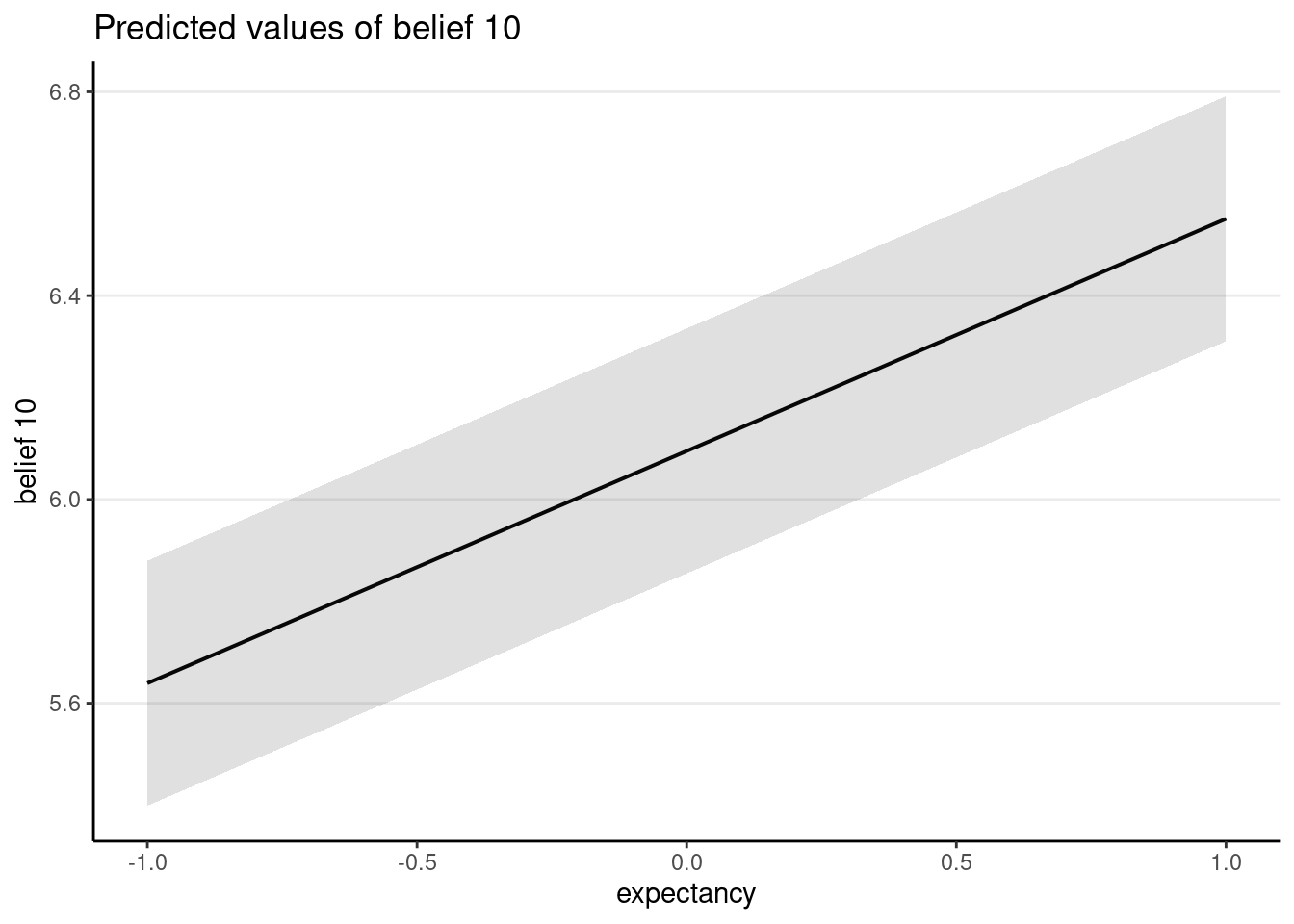
Mediator → Outcome, Holding Treatment Constant
Because the mediator has a cluster mean component, we should also include it.
dat <- dat |>
group_by(sub) |>
mutate(belief10_pm = mean(belief10)) |>
ungroup()# Component (b). Make sure to include the treatment variable.
mb <- lmer(jol10 ~ belief10 + belief10_pm + expectancy +
(belief10 | sub), data = dat)
plot_model(mb, type = "pred", terms = "belief10")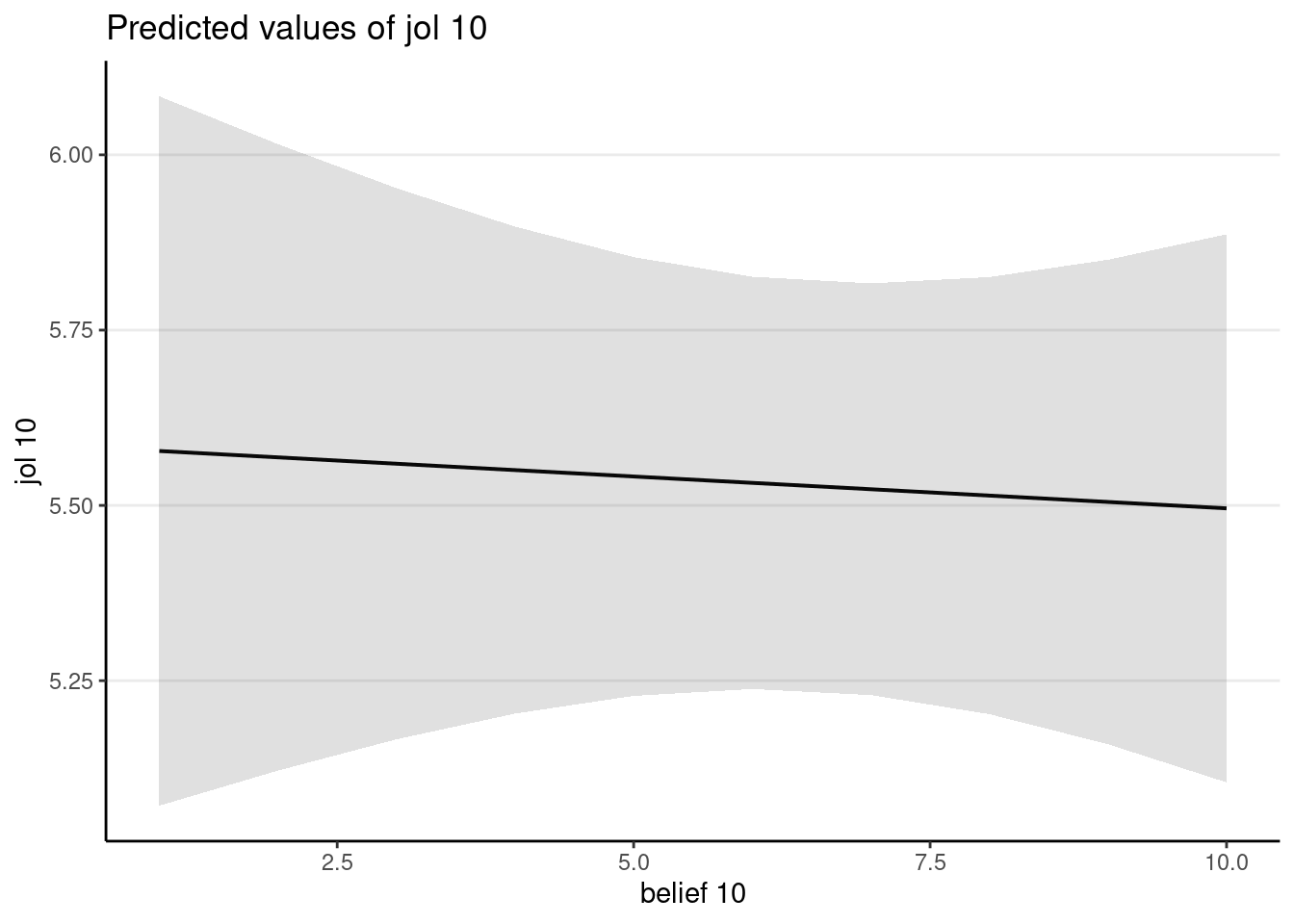
The mediator does not have a strong direct effect on the outcome.
Mediation Analysis
You should only perform a mediation analysis when the data support the causal directions of the variables, or when you are willing to make strong causal assumptions. You can use the mediation::mediate() function.
# Input: components (a) and (b). This takes a few minutes because
# it uses simulations.
m_med <- mediate(
ma,
model.y = mb,
treat = "expectancy", # name of mediator
mediator = "belief10", # name of mediator
control.value = -1,
treat.value = 1
)
summary(m_med)>#
># Causal Mediation Analysis
>#
># Quasi-Bayesian Confidence Intervals
>#
># Mediator Groups: sub
>#
># Outcome Groups: sub
>#
># Output Based on Overall Averages Across Groups
>#
># Estimate 95% CI Lower 95% CI Upper p-value
># ACME -0.0056 -0.0878 0.07 0.88
># ADE 0.5677 0.4536 0.67 <2e-16 ***
># Total Effect 0.5621 0.4606 0.67 <2e-16 ***
># Prop. Mediated -0.0097 -0.1658 0.12 0.88
># ---
># Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
>#
># Sample Size Used: 7680
>#
>#
># Simulations: 1000plot(m_med)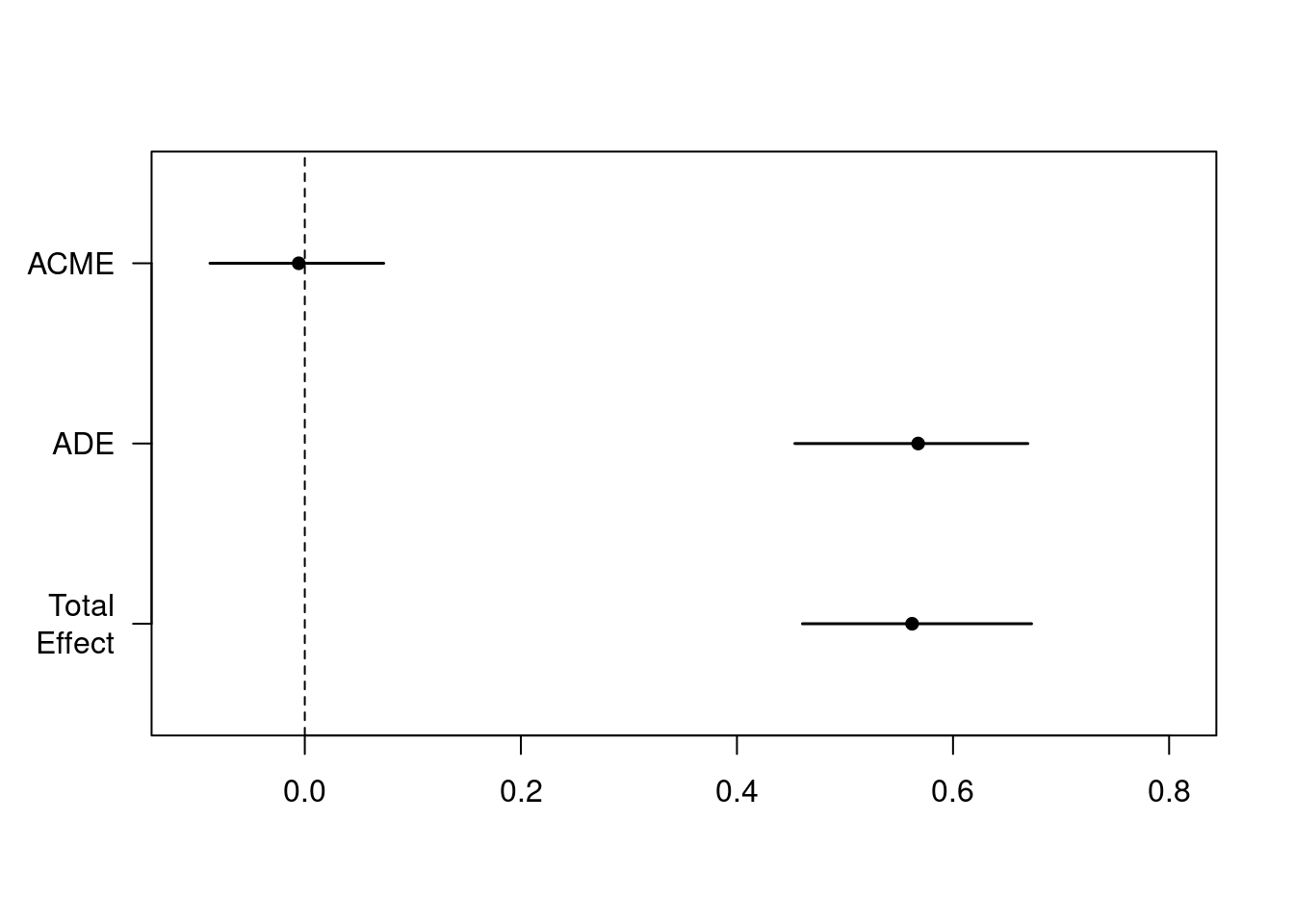
The row labeled ACME is the estimated mediation effect, which is not statistically significant in this example.