# To install a package, run the following ONCE (and only once on your computer)
# install.packages("psych")
library(here) # makes reading data more consistent
library(tidyverse) # for data manipulation and plotting
library(haven) # for importing SPSS/SAS/Stata data
library(lme4) # for multilevel analysis
library(sjPlot) # for plotting effects
library(MuMIn) # for computing r-squared
library(broom.mixed) # for augmenting results
library(modelsummary) # for making tables
library(interactions) # for plotting interactionsWeek 4 (R): Lv-1 Predictors
Load Packages and Import Data
Import Data
# Read in the data (pay attention to the directory)
hsball <- read_sav(here("data_files", "hsball.sav"))Between-Within Decomposition
Cluster-mean centering
To separate the effects of a lv-1 predictor into different levels, one needs to first center the predictor on the cluster means:
hsball <- hsball %>%
group_by(id) %>% # operate within schools
mutate(ses_cm = mean(ses), # create cluster means (the same as `meanses`)
ses_cmc = ses - ses_cm) %>% # cluster-mean centered
ungroup() # exit the "editing within groups" mode
# The meanses variable already exists in the original data, but it's slightly
# different from computing it by hand
hsball %>%
select(id, ses, meanses, ses_cm, ses_cmc)id <chr> | ses <dbl> | meanses <dbl> | ses_cm <dbl> | ses_cmc <dbl> |
|---|---|---|---|---|
| 1224 | -1.528 | -0.428 | -0.434382979 | -1.093617e+00 |
| 1224 | -0.588 | -0.428 | -0.434382979 | -1.536170e-01 |
| 1224 | -0.528 | -0.428 | -0.434382979 | -9.361702e-02 |
| 1224 | -0.668 | -0.428 | -0.434382979 | -2.336170e-01 |
| 1224 | -0.158 | -0.428 | -0.434382979 | 2.763830e-01 |
| 1224 | 0.022 | -0.428 | -0.434382979 | 4.563830e-01 |
| 1224 | -0.618 | -0.428 | -0.434382979 | -1.836170e-01 |
| 1224 | -0.998 | -0.428 | -0.434382979 | -5.636170e-01 |
| 1224 | -0.888 | -0.428 | -0.434382979 | -4.536170e-01 |
| 1224 | -0.458 | -0.428 | -0.434382979 | -2.361702e-02 |
Model Equation
Lv-1:
Lv-2:
sesrepresenting the expected difference in student achievement between two students in the same school with one unit difference inses,
meanses.
Running in R
We can specify the model as:
m_bw <- lmer(mathach ~ meanses + ses_cmc + (1 | id), data = hsball)As an alternative to creating the cluster means and centered variables in the data, you can directly create in the formula:
m_bw2 <- lmer(mathach ~ I(ave(ses, id)) + I(ses - ave(ses, id)) + (1 | id),
data = hsball)You can summarize the results using
summary(m_bw)># Linear mixed model fit by REML ['lmerMod']
># Formula: mathach ~ meanses + ses_cmc + (1 | id)
># Data: hsball
>#
># REML criterion at convergence: 46568.6
>#
># Scaled residuals:
># Min 1Q Median 3Q Max
># -3.1666 -0.7253 0.0174 0.7557 2.9453
>#
># Random effects:
># Groups Name Variance Std.Dev.
># id (Intercept) 2.692 1.641
># Residual 37.019 6.084
># Number of obs: 7185, groups: id, 160
>#
># Fixed effects:
># Estimate Std. Error t value
># (Intercept) 12.6481 0.1494 84.68
># meanses 5.8662 0.3617 16.22
># ses_cmc 2.1912 0.1087 20.16
>#
># Correlation of Fixed Effects:
># (Intr) meanss
># meanses -0.004
># ses_cmc 0.000 0.000Contextual Effect
With a level-1 predictor like ses, which has both student-level and school-level variance, we can include both the level-1 and the school mean variables as predictors. When the level-1 predictor is present, the coefficient for the group means variable becomes the contextual effect.
Model Equation
Lv-1:
Lv-2:
sesrepresenting the expected difference in student achievement between two students in the same school with one unit difference inses,
sesbut from two schools with one unit difference inmeanses.
Running in R
We can specify the model as:
contextual <- lmer(mathach ~ meanses + ses + (1 | id), data = hsball)You can summarize the results using
summary(contextual)># Linear mixed model fit by REML ['lmerMod']
># Formula: mathach ~ meanses + ses + (1 | id)
># Data: hsball
>#
># REML criterion at convergence: 46568.6
>#
># Scaled residuals:
># Min 1Q Median 3Q Max
># -3.1666 -0.7253 0.0174 0.7557 2.9453
>#
># Random effects:
># Groups Name Variance Std.Dev.
># id (Intercept) 2.692 1.641
># Residual 37.019 6.084
># Number of obs: 7185, groups: id, 160
>#
># Fixed effects:
># Estimate Std. Error t value
># (Intercept) 12.6613 0.1494 84.763
># meanses 3.6750 0.3777 9.731
># ses 2.1912 0.1087 20.164
>#
># Correlation of Fixed Effects:
># (Intr) meanss
># meanses -0.005
># ses 0.004 -0.288If you compare the REML criterion at convergence number, you can see this is the same as the between-within model. The estimated contextual effect is the coefficient of meanses minus the coefficient of ses_cmc, which is the same as what you will get in the contextual effect model.
Random-Coefficients Model
Explore Variations in Slopes
hsball %>%
# randomly sample 16 schools
filter(id %in% sample(unique(id), 16)) %>%
# ses on x-axis and mathach on y-axis
ggplot(aes(x = ses, y = mathach)) +
# add points (half the original size)
geom_point(size = 0.5) +
# add a linear smoother
geom_smooth(method = "lm") +
# present data of different ids in different panels
facet_wrap(~id)># `geom_smooth()` using formula = 'y ~ x'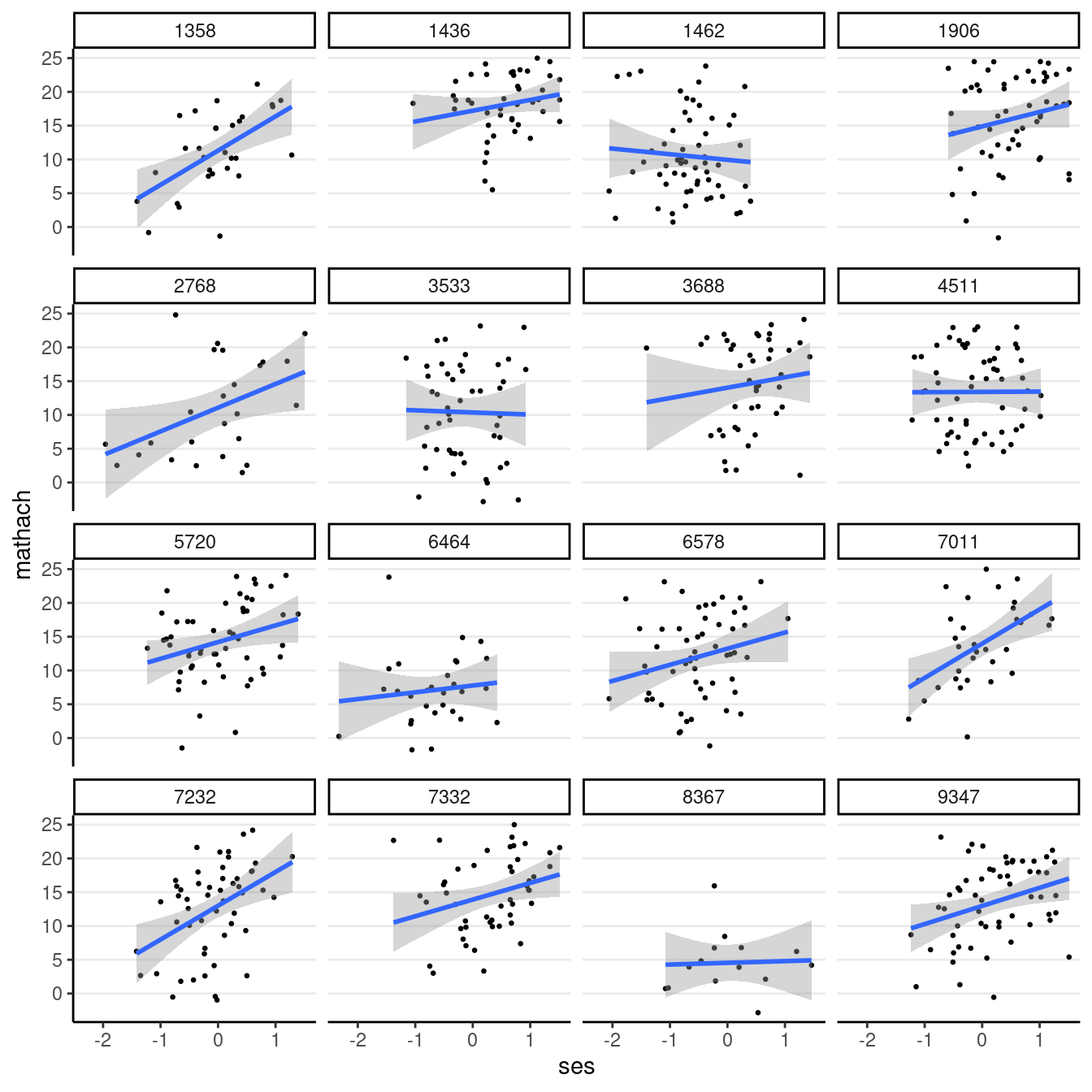
Model Equation
Lv-1:
Lv-2:
Running in R
We have to put ses in the random part:
ran_slp <- lmer(mathach ~ meanses + ses_cmc + (ses_cmc | id), data = hsball)You can summarize the results using
summary(ran_slp)># Linear mixed model fit by REML ['lmerMod']
># Formula: mathach ~ meanses + ses_cmc + (ses_cmc | id)
># Data: hsball
>#
># REML criterion at convergence: 46557.7
>#
># Scaled residuals:
># Min 1Q Median 3Q Max
># -3.1768 -0.7269 0.0160 0.7560 2.9406
>#
># Random effects:
># Groups Name Variance Std.Dev. Corr
># id (Intercept) 2.6931 1.6411
># ses_cmc 0.6858 0.8281 -0.19
># Residual 36.7132 6.0591
># Number of obs: 7185, groups: id, 160
>#
># Fixed effects:
># Estimate Std. Error t value
># (Intercept) 12.6454 0.1492 84.74
># meanses 5.8963 0.3600 16.38
># ses_cmc 2.1913 0.1280 17.11
>#
># Correlation of Fixed Effects:
># (Intr) meanss
># meanses -0.004
># ses_cmc -0.088 0.004Showing the random intercepts and slopes
head(coef(ran_slp)$id) # `head()` shows only the first six schools(Intercept) <dbl> | meanses <dbl> | ses_cmc <dbl> | ||
|---|---|---|---|---|
| 1224 | 12.32411 | 5.896293 | 2.292981 | |
| 1288 | 12.69256 | 5.896293 | 2.359732 | |
| 1296 | 10.70293 | 5.896293 | 2.040818 | |
| 1308 | 12.94395 | 5.896293 | 2.013578 | |
| 1317 | 11.46711 | 5.896293 | 2.092983 | |
| 1358 | 11.64868 | 5.896293 | 2.800057 |
Plotting Random Slopes
# The `augment` function adds predicted values to the data
augment(ran_slp, data = hsball) %>%
ggplot(aes(x = ses, y = .fitted, color = factor(id))) +
geom_smooth(method = "lm", se = FALSE, size = 0.5) +
labs(y = "Predicted mathach") +
guides(color = "none")># Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
># ℹ Please use `linewidth` instead.># `geom_smooth()` using formula = 'y ~ x'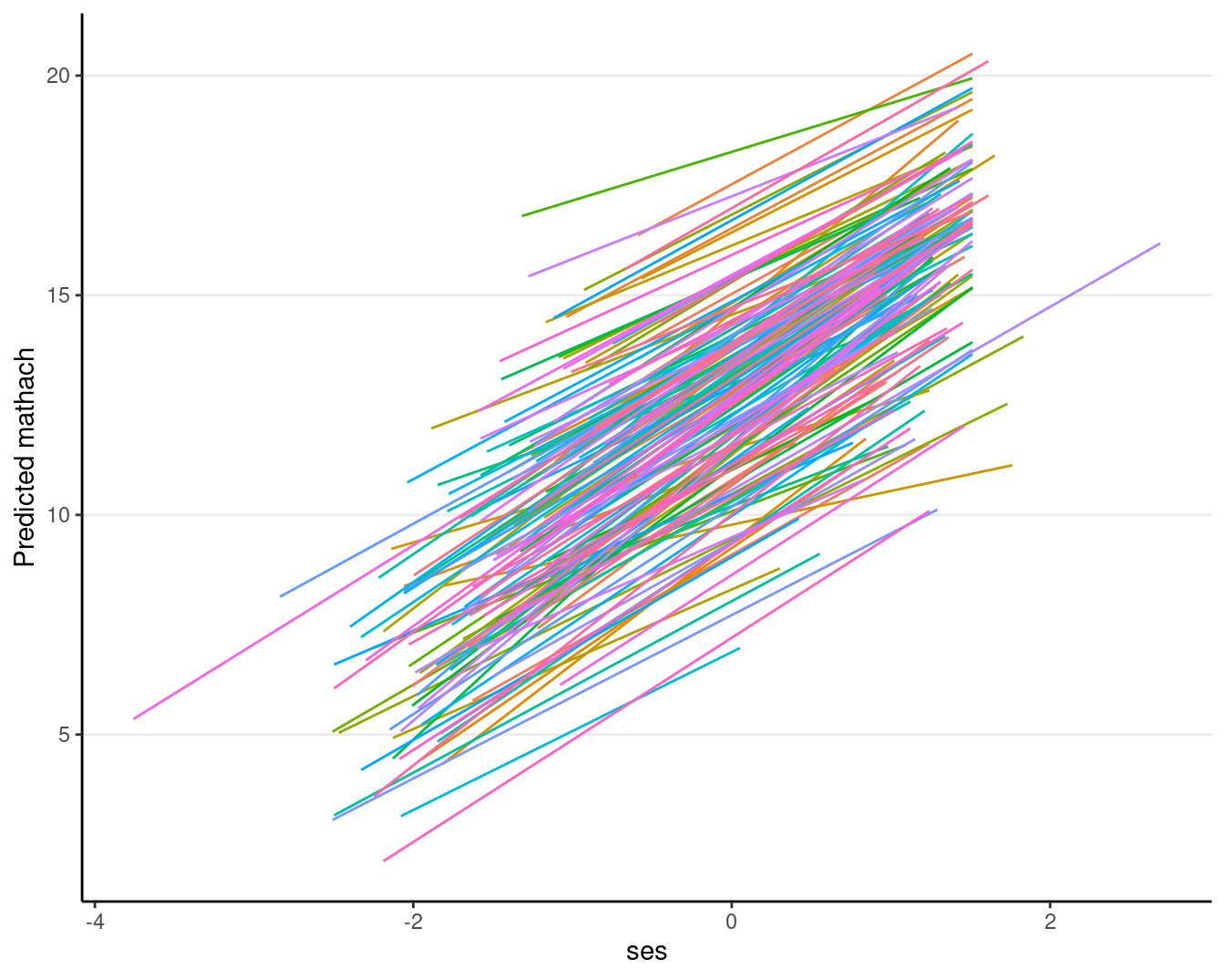
Or using sjPlot::plot_model() (with ses_cmc, not ses, on the x-axis)
plot_model(ran_slp, type = "pred", pred.type = "re", terms = c("ses_cmc", "id"),
# suppress confidence band
ci.lvl = NA, show.legend = FALSE, colors = "gs")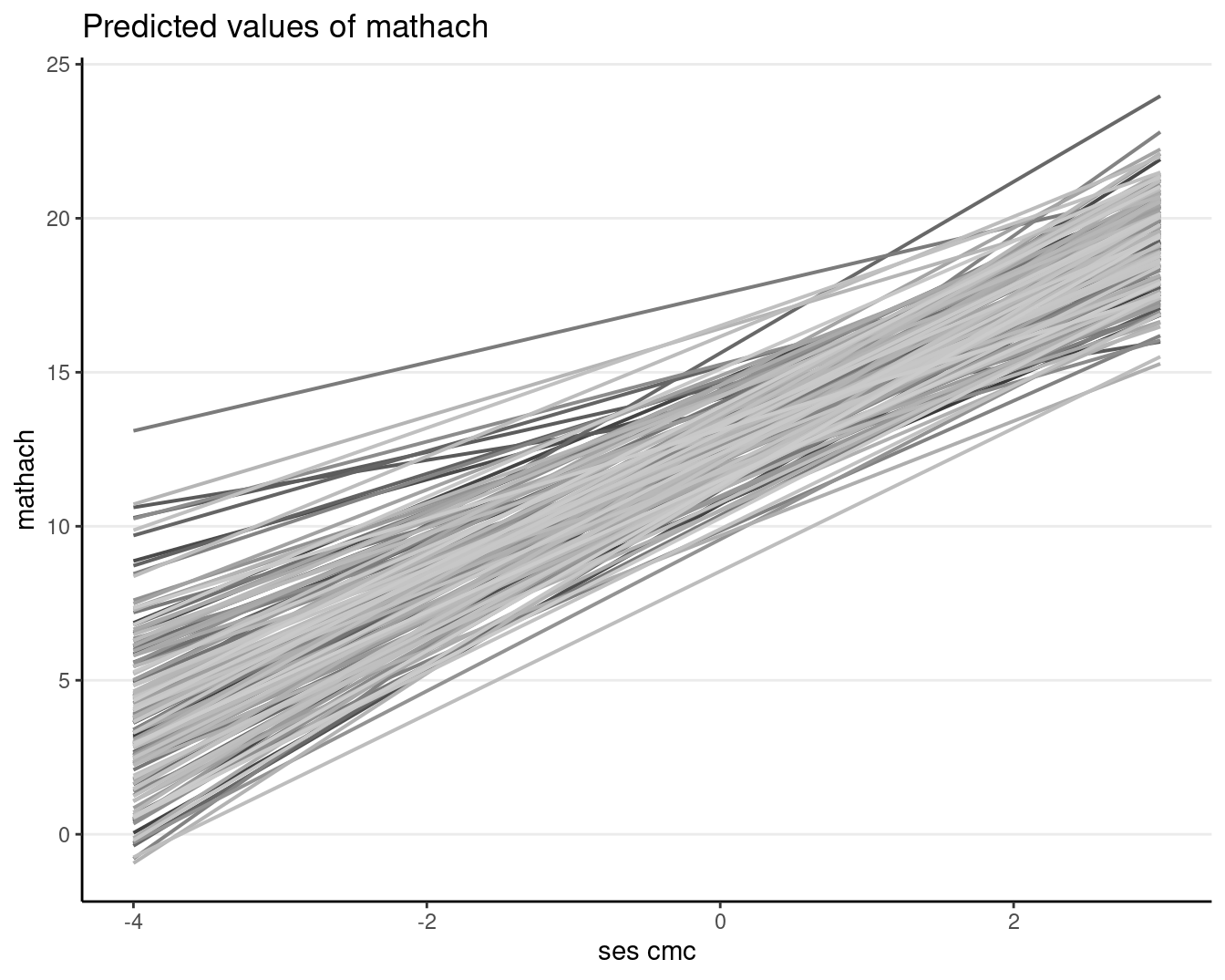
Including the effect of meanses
# augmented data (adding EB estimates)
augment(ran_slp, data = hsball) %>%
ggplot(aes(x = ses, y = mathach, color = factor(id))) +
# Add points
geom_point(size = 0.2, alpha = 0.2) +
# Add within-cluster lines
geom_smooth(aes(y = .fitted),
method = "lm", se = FALSE, size = 0.5) +
# Add group means with `stat_summary`
stat_summary(aes(x = meanses, y = .fitted,
fill = factor(id)),
fun = mean,
geom = "point",
color = "red", # add border
shape = 24, # use triangles
size = 2.5) +
# Add between coefficient
geom_smooth(aes(x = meanses, y = .fitted),
method = "lm", se = FALSE,
color = "black") +
labs(y = "Math Achievement") +
# Suppress legend as there are too many schools
guides(color = "none", fill = "none")># `geom_smooth()` using formula = 'y ~ x'
># `geom_smooth()` using formula = 'y ~ x'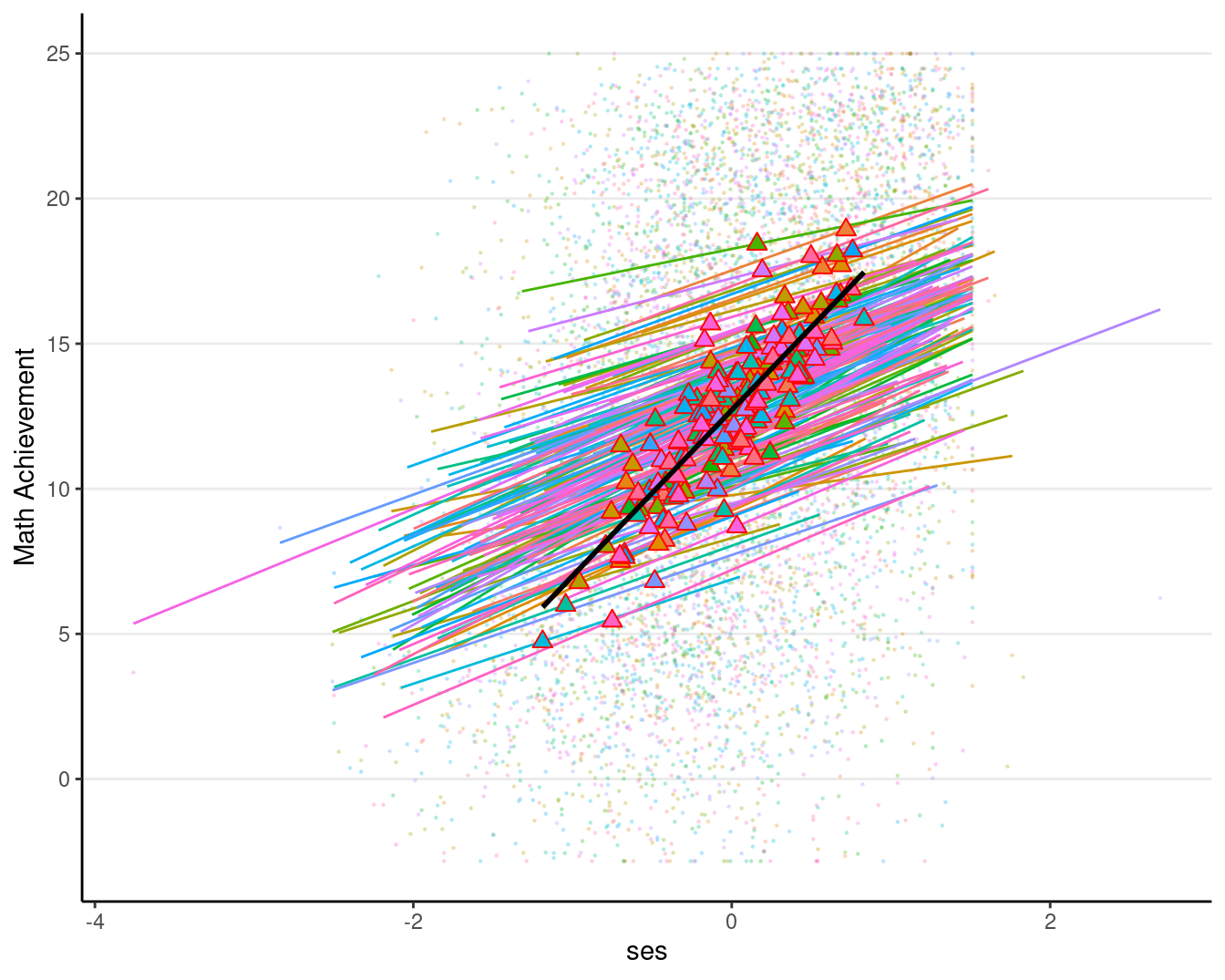
Or on separate graphs:
# Create a common base graph
pbase <- augment(ran_slp, data = hsball) %>%
ggplot(aes(x = ses, y = mathach, color = factor(id))) +
# Add points
geom_point(size = 0.2, alpha = 0.2) +
labs(y = "Math Achievement") +
# Suppress legend
guides(color = "none")
# Lv-1 effect
p1 <- pbase +
# Add within-cluster lines
geom_smooth(aes(y = .fitted),
method = "lm", se = FALSE, size = 0.5)
# Lv-2 effect
p2 <- pbase +
# Add group means
stat_summary(aes(x = meanses, y = .fitted),
fun = mean,
geom = "point",
shape = 17, # use triangles
size = 2.5) +
# Add between coefficient
geom_smooth(aes(x = meanses, y = .fitted),
method = "lm", se = FALSE,
color = "black")
# Put the two graphs together (need the gridExtra package)
gridExtra::grid.arrange(p1, p2, ncol = 2) # in two columns># `geom_smooth()` using formula = 'y ~ x'
># `geom_smooth()` using formula = 'y ~ x'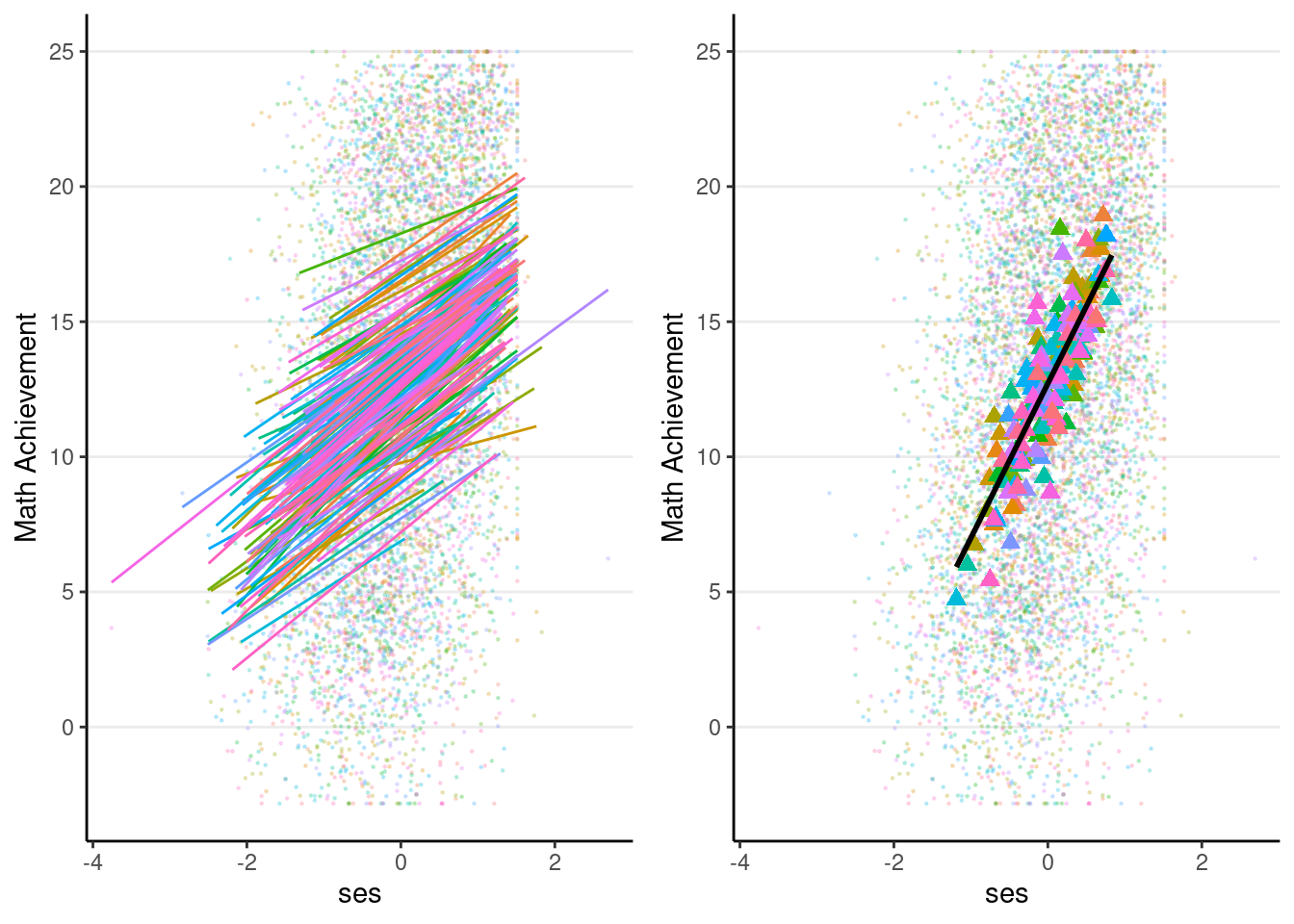
Effect Size
As discussed in the previous week, we can obtain the effect size (
(r2_ran_slp <- r.squaredGLMM(ran_slp))># Warning: 'r.squaredGLMM' now calculates a revised statistic. See the help page.># R2m R2c
># [1,] 0.1684161 0.2310862Analyzing Cross-Level Interactions
sector is added to the level-2 intercept and slope equations.
Model Equation
Lv-1:
Lv-2:
sector variable representing the expected difference in achievement between two students with the same SES level and from two schools with the same school-level SES, but one is a catholic school and the other a private school. -
Running in R
We have to put the sector * ses interaction to the fixed part:
# The first step is optional, but let's recode sector into a factor variable
hsball <- hsball %>%
mutate(sector = factor(sector, levels = c(0, 1),
labels = c("Public", "Catholic")))
crlv_int <- lmer(mathach ~ meanses + sector * ses_cmc + (ses_cmc | id),
data = hsball)You can summarize the results using
summary(crlv_int)># Linear mixed model fit by REML ['lmerMod']
># Formula: mathach ~ meanses + sector * ses_cmc + (ses_cmc | id)
># Data: hsball
>#
># REML criterion at convergence: 46514.5
>#
># Scaled residuals:
># Min 1Q Median 3Q Max
># -3.11560 -0.72905 0.01592 0.75280 3.01543
>#
># Random effects:
># Groups Name Variance Std.Dev. Corr
># id (Intercept) 2.3807 1.5430
># ses_cmc 0.2716 0.5212 0.24
># Residual 36.7077 6.0587
># Number of obs: 7185, groups: id, 160
>#
># Fixed effects:
># Estimate Std. Error t value
># (Intercept) 12.0846 0.1987 60.81
># meanses 5.2450 0.3682 14.24
># sectorCatholic 1.2523 0.3062 4.09
># ses_cmc 2.7877 0.1559 17.89
># sectorCatholic:ses_cmc -1.3478 0.2348 -5.74
>#
># Correlation of Fixed Effects:
># (Intr) meanss sctrCt ss_cmc
># meanses 0.245
># sectorCthlc -0.697 -0.355
># ses_cmc 0.071 -0.002 -0.046
># sctrCthlc:_ -0.048 -0.001 0.071 -0.664Effect Size
By comparing the
(r2_crlv_int <- r.squaredGLMM(crlv_int))># R2m R2c
># [1,] 0.1759122 0.2284431# Change
r2_crlv_int - r2_ran_slp># R2m R2c
># [1,] 0.007496143 -0.002643047Therefore, the cross-level interaction between sector and SES accounted for an effect size of
Simple Slopes
With a cross-level interaction, the slope between ses_cmc and mathach depends on a moderator, sector. To find out what the model suggests to be the slope at a particular level of sector, which is also called a simple slope in the literature, you just need to look at the equation for sector = 0 (public school), the simple slope is sector = 1 (private school), the simple slope is
Plotting the Interactions
crlv_int %>%
augment(data = hsball) %>%
ggplot(aes(
x = ses, y = .fitted, group = factor(id),
color = factor(sector) # use `sector` for coloring lines
)) +
geom_smooth(method = "lm", se = FALSE, size = 0.5) +
labs(y = "Predicted mathach", color = "sector")># `geom_smooth()` using formula = 'y ~ x'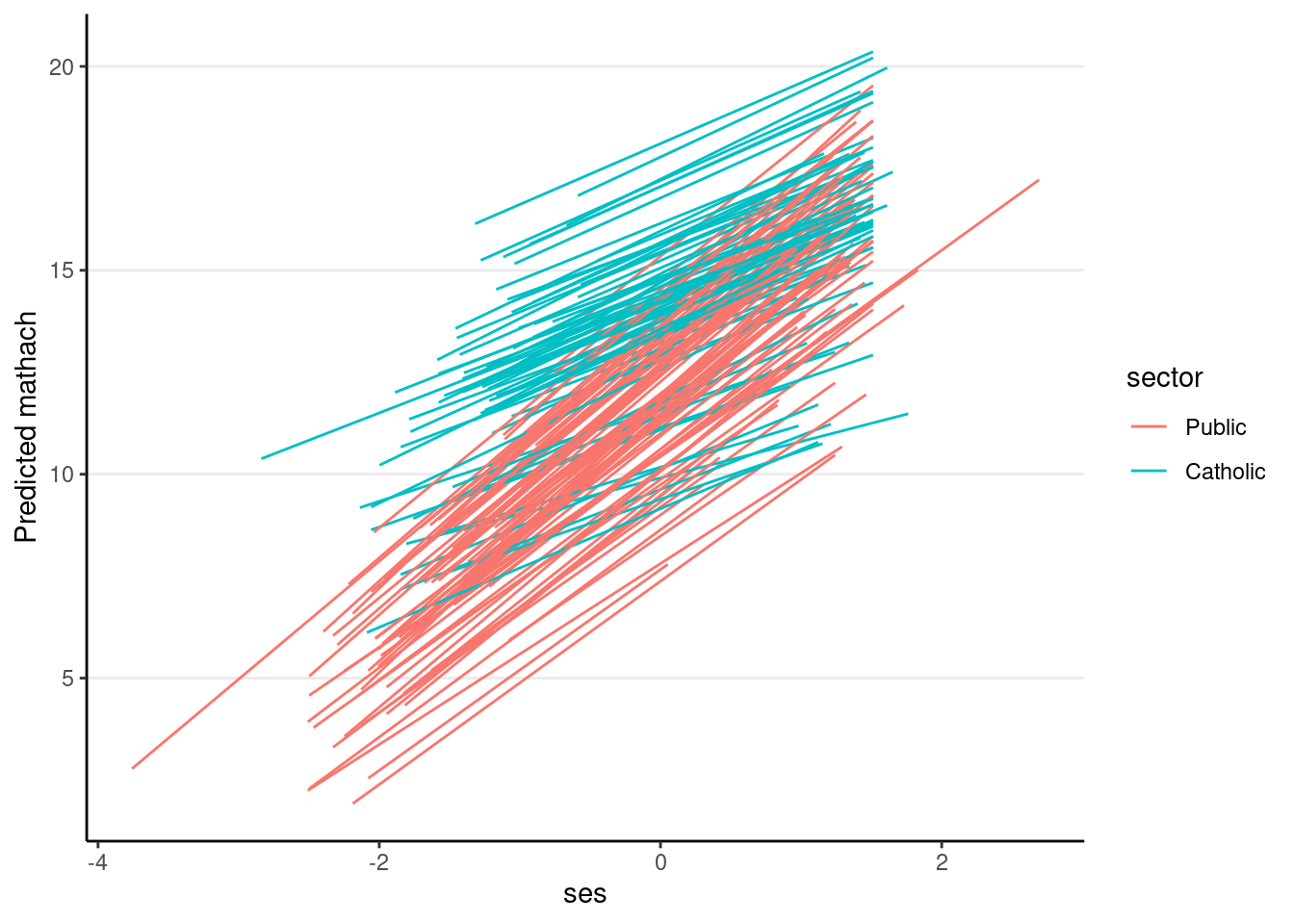
You can also use the sjPlot::plot_model() function for just the fixed effects:
plot_model(crlv_int, type = "pred", pred.type = "re",
terms = c("ses_cmc", "id", "sector"),
# suppress confidence band
ci.lvl = NA, show.legend = FALSE, colors = "gs",
# suppress title
title = "",
axis.title = c("SES (cluster-mean centered)", "Math Achievement"),
show.data = TRUE, dot.size = 0.5, dot.alpha = 0.5)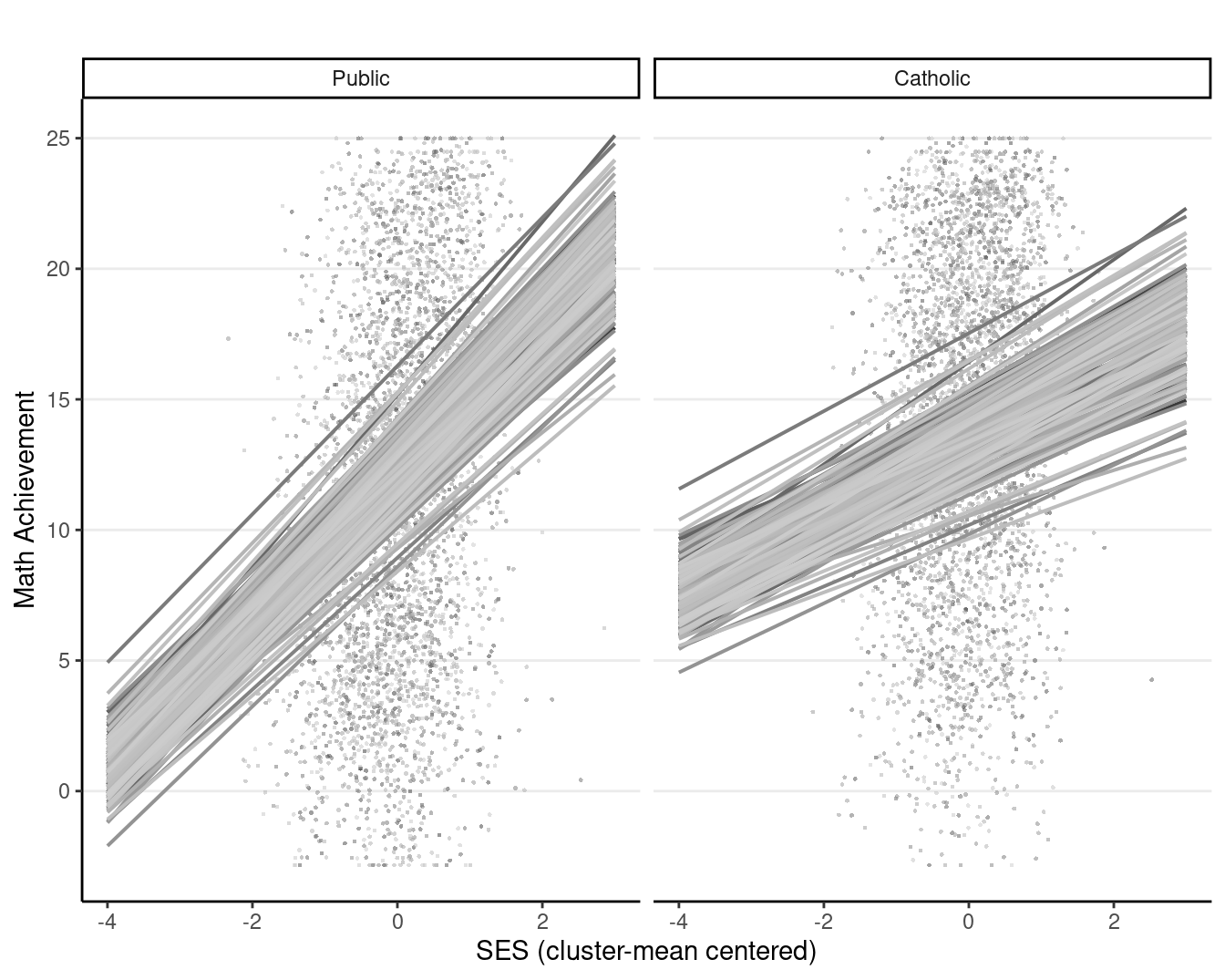
Summaizing Analyses in a Table
You can again use the msummary() function from the modelsummary package to quickly summarize the results in a table.
# Basic table from `modelsummary`
msummary(list(
"Between-within" = m_bw,
"Contextual" = contextual,
"Random slope" = ran_slp,
"Cross-level interaction" = crlv_int
))| Between-within | Contextual | Random slope | Cross-level interaction | |
|---|---|---|---|---|
| (Intercept) | 12.648 | 12.661 | 12.645 | 12.085 |
| (0.149) | (0.149) | (0.149) | (0.199) | |
| meanses | 5.866 | 3.675 | 5.896 | 5.245 |
| (0.362) | (0.378) | (0.360) | (0.368) | |
| ses_cmc | 2.191 | 2.191 | 2.788 | |
| (0.109) | (0.128) | (0.156) | ||
| ses | 2.191 | |||
| (0.109) | ||||
| sectorCatholic | 1.252 | |||
| (0.306) | ||||
| sectorCatholic × ses_cmc | −1.348 | |||
| (0.235) | ||||
| SD (Intercept id) | 1.641 | 1.641 | 1.641 | 1.543 |
| SD (ses_cmc id) | 0.828 | 0.521 | ||
| Cor (Intercept~ses_cmc id) | −0.195 | 0.244 | ||
| SD (Observations) | 6.084 | 6.084 | 6.059 | 6.059 |
| Num.Obs. | 7185 | 7185 | 7185 | 7185 |
| R2 Marg. | 0.167 | 0.167 | 0.168 | 0.176 |
| R2 Cond. | 0.224 | 0.224 | 0.231 | 0.228 |
| AIC | 46578.6 | 46578.6 | 46571.7 | 46532.5 |
| BIC | 46613.0 | 46613.0 | 46619.8 | 46594.4 |
| ICC | 0.07 | 0.07 | 0.08 | 0.06 |
| RMSE | 6.03 | 6.03 | 5.99 | 6.00 |
Bonus: More “APA-like” Table
Make it look like the table at https://apastyle.apa.org/style-grammar-guidelines/tables-figures/sample-tables#regression. You can see a recent recommendation in this paper published in the Journal of Memory and Language
# Step 1. Create a map for the predictors and the terms in the model.
# Need to use '\\( \\)' to show math.
# Rename and reorder the rows. Need to use '\\( \\)' to
# show math. If this does not work for you, don't worry about it.
cm <- c("(Intercept)" = "Intercept",
"meanses" = "school mean SES",
"ses_cmc" = "SES (cluster-mean centered)",
"sectorCatholic" = "Catholic school",
"sectorCatholicses_cmc" = "Catholic school x SES (cmc)",
"SD (Intercept id)" = "\\(\\tau_0\\)",
"SD (ses_cmc id)" = "\\(\\tau_1\\) (SES)",
"Cor (Intercept~ses_cmc id)" = "\\(\\tau_{01}\\)",
"SD (Observations)" = "\\(\\sigma\\)")
# Step 2. Create a list of models to have one column for coef,
# one for SE, one for CI, and one for p
models <- list(
"Estimate" = crlv_int,
"SE" = crlv_int,
"95% CI" = crlv_int
)
# Step 3. Add rows to say fixed and random effects
# (Need same number of columns as the table)
new_rows <- data.frame(
term = c("Fixed effects", "Random effects"),
est = c("", ""),
se = c("", ""),
ci = c("", "")
)
# Specify which rows to add
attr(new_rows, "position") <- c(1, 7)
# Step 4. Render the table
msummary(models,
estimate = c("estimate", "std.error",
"[{conf.low}, {conf.high}]"),
statistic = NULL, # suppress the extra rows for SEs
# suppress gof indices (e.g., R^2)
gof_omit = ".*",
coef_map = cm,
add_rows = new_rows)| Estimate | SE | 95% CI | |
|---|---|---|---|
| Fixed effects | |||
| Intercept | 12.085 | 0.199 | [11.695, 12.474] |
| school mean SES | 5.245 | 0.368 | [4.523, 5.967] |
| SES (cluster-mean centered) | 2.788 | 0.156 | [2.482, 3.093] |
| Catholic school | 1.252 | 0.306 | [0.652, 1.853] |
| |
1.543 | ||
| Random effects | |||
| |
0.521 | ||
| |
0.244 | ||
| |
6.059 |