# To install a package, run the following ONCE (and only once on your computer)
# install.packages("psych")
library(here) # makes reading data more consistent
library(tidyverse) # for data manipulation and plotting
library(haven) # for importing SPSS/SAS/Stata data
library(lme4) # for multilevel logistic models
library(brms) # for Bayesian multilevel models
library(sjPlot) # for plotting
library(MuMIn) # for R^2
library(modelsummary) # for making tablesWeek 12 (R): Logistic
Load Packages and Import Data
This week, we’ll talk about logistic regression for binary data. We’ll use the same HSB data you have seen before, but with a dichotomous variable created. In practice, it is generally a bad idea to arbitrarily dichotomize a binary variable, as it may represent a substantial loss of information; it’s done here just for pedagogical purpose.
Here we consider those who score 20 or above for a “commended” status.
# Read in the data (pay attention to the directory)
hsball <- read_sav(here("data_files", "hsball.sav"))
# Dichotomize mathach
hsball <- hsball %>%
mutate(mathcom = as.integer(mathach >= 20))Let’s plot the commended distribution for a few schools.
set.seed(7351)
# Randomly select some schools
random_schools <- sample(hsball$id, size = 9)
hsball %>%
filter(id %in% random_schools) %>%
mutate(mathcom = factor(mathcom,
labels = c("not commended", "commended")
)) %>%
ggplot(aes(x = mathcom)) +
geom_bar() +
facet_wrap(~id, ncol = 3) +
coord_flip()
Problem of a Linear Model
Let’s use our linear model, and then talk about the problems. Consider first meanses as a predictor.
m_lme <- lmer(mathcom ~ meanses + (1 | id), data = hsball)
summary(m_lme)># Linear mixed model fit by REML ['lmerMod']
># Formula: mathcom ~ meanses + (1 | id)
># Data: hsball
>#
># REML criterion at convergence: 6258.7
>#
># Scaled residuals:
># Min 1Q Median 3Q Max
># -1.1991 -0.5290 -0.3941 -0.1301 2.6181
>#
># Random effects:
># Groups Name Variance Std.Dev.
># id (Intercept) 0.005148 0.07175
># Residual 0.136664 0.36968
># Number of obs: 7185, groups: id, 160
>#
># Fixed effects:
># Estimate Std. Error t value
># (Intercept) 0.178404 0.007222 24.70
># meanses 0.178190 0.017482 10.19
>#
># Correlation of Fixed Effects:
># (Intr)
># meanses -0.006So it runs and indicates a positive association between meanses and mathcom. Let’s plot the effect:
plot_model(m_lme,
type = "pred", terms = "meanses",
show.data = TRUE, jitter = 0.02,
title = "", dot.size = 0.5
)
Well, this looks a bit strange. The outcome can only take two values, but the predictions have decimals. We can assume maybe we are predicting the probability of being commended for each student, but still, there are some impossible negative predictions. For example, if we consider a school with meanses = -2, our prediction would be
predict(m_lme, newdata = data.frame(meanses = -2, id = NA), re.form = NA)># 1
># -0.1779765Another problem is that the data is not normally distributed. We can look at the distribution of the data (in a histogram) and simulated data based on the model (in lighter lines) below:
# Simulate a data set based on the linear model
sim_y <- simulate(m_lme)
ggplot(hsball, aes(x = mathcom)) +
geom_histogram(aes(y = ..density..)) +
geom_density(aes(x = sim_y[, 1]), alpha = 0.5)># Warning: The dot-dot notation (`..density..`) was deprecated in ggplot2 3.4.0.
># ℹ Please use `after_stat(density)` instead.># `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
Another related and subtle problem is that the residual variance is not constant across levels of the predictor. For example, the residual variance for meanses
broom.mixed::augment(m_lme) %>%
filter(meanses <= -.5) %>%
# residual variance
summarize(`var(e | meanses <= .5)` = var(.resid))var(e | meanses <= .5) <dbl> | |||
|---|---|---|---|
| 0.05823345 |
But when `meanses > 0.5, it was
broom.mixed::augment(m_lme) %>%
filter(meanses > -.5) %>%
# residual variance
summarize(`var(e | meanses > .5)` = var(.resid))var(e | meanses > .5) <dbl> | |||
|---|---|---|---|
| 0.1453736 |
These are pretty large differences, and it happens every time one uses a normal linear model for a binary outcome. The textbook (section 17.1) has more discussion on this.
So because of these limitations, we generally prefer models designed for binary outcomes, the most popular one is commonly referred to as a logistic model.
Multilevel Logistic Regression
The logistic model modifies the linear multilevel model in two ways.
Bernoulli Distribution
First, it replaces the assumption that the conditional distribution of the outcome is normal with one that says the outcome follows a Bernoulli distribution, which is a distribution for binary variables (e.g., the outcome of a coin flip). Below is a comparison of a Bernoulli distribution (left) with mean = 0.8 (i.e., 80% success and 20% failure), which by definition has a standard deviation of 0.4, and a normal distribution (right) with a mean = 0.8 and a standard deviation of 0.4:
p1 <- ggplot(tibble(y = c(0, 1), p = c(0.2, 0.8)), aes(x = y, y = p)) +
geom_col(width = 0.05) +
xlim(-0.4, 2) +
labs(y = "Probability Mass")
p2 <- ggplot(tibble(y = c(0, 1)), aes(x = y)) +
stat_function(fun = dnorm, args = list(mean = 0.8, sd = 0.4)) +
xlim(-0.4, 2) +
labs(y = "Probability Density")
gridExtra::grid.arrange(p1, p2, ncol = 2)
With the Bernoulli distribution, the possible values are only 0 and 1, so it matches the outcome.
Transformation
Second, instead of directly modeling the mean of a binary outcome (i.e., probability), which is bounded between 0 and 1, a logistic model transforms the mean of the outcome into something unbounded (i.e., with a range between
As shown below,
ggplot(tibble(mu = c(0, 1)), aes(x = mu)) +
stat_function(fun = qlogis, n = 501) +
labs(x = "Probability", y = "Log Odds (logit)")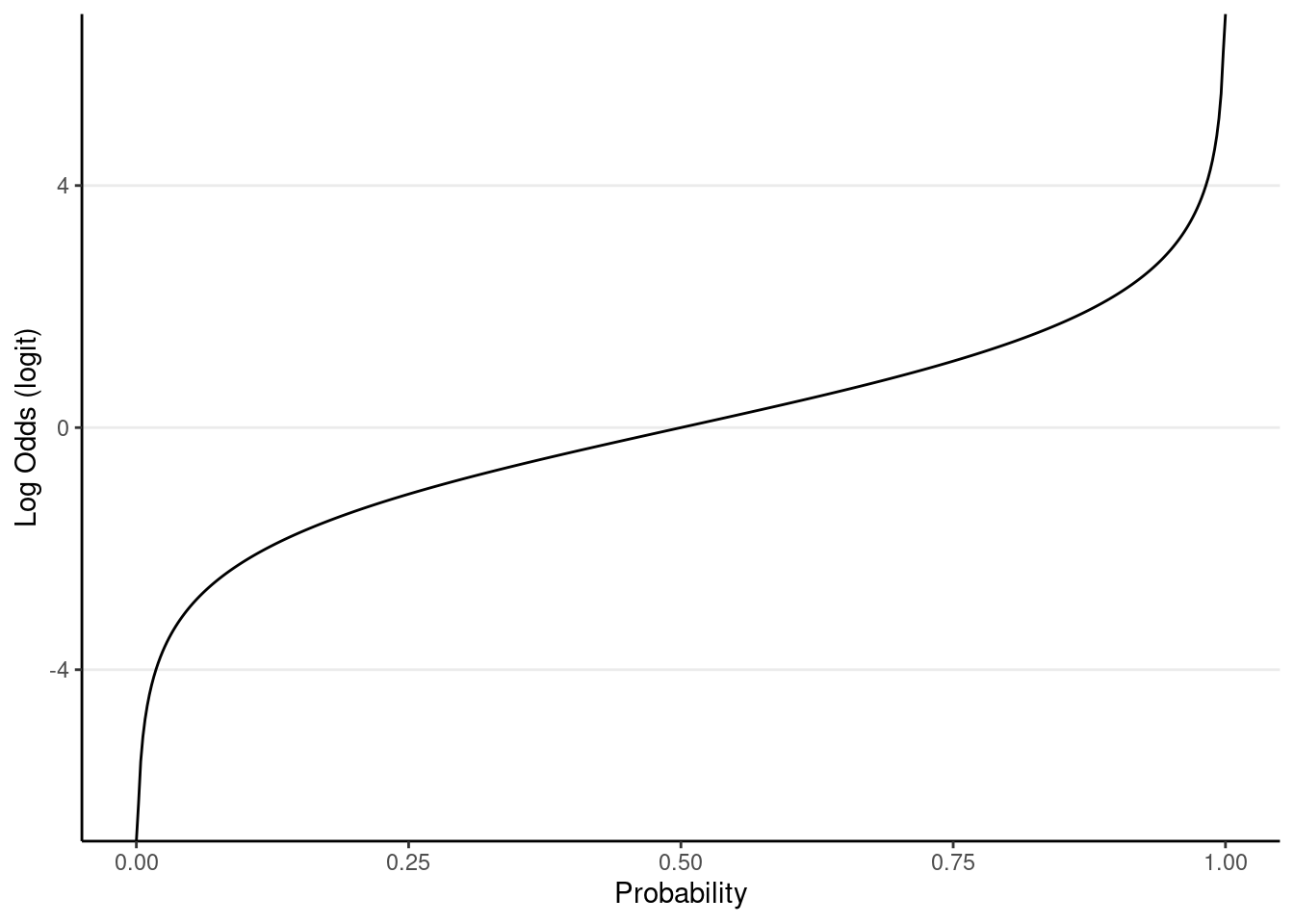
For example, when the probability is 0.5, the log odds = 0; when the probability = 0.9, the log odds = 0.7109495; when the probability = -0.9, the log odds = 0.2890505.
Model Equation
To understand the equation for a logistic model, it helps to write the normal linear model in its full form by specifying the distributions. Let’s look at the simplest unconditional model without predictors. First, we can write mathcom for the mathcom is normally distributed around the predicted value with an error variance of
Now, we’ll change the distribution of mathcom, and transform
Level 1
Using brms
With logistic regression, frequentist methods rely on approximations. Sometimes they may be problematic, especially for models with random slopes, so attention needs to be paid to assess convergence. Compared to linear models, the only thing you need to change is to specify family = binomial("logit") (for brms) or family = bernoulli("logit") (for lme4).
Unconditional Model
Note: Maximum likelihood estimation with logistic models is more challenging, as the likelihood functions involve integrals that are difficult to solve. Different software/packages may use different algorithms to solve that, and they have different degrees of accuracy.
m0_logit <- glmer(mathcom ~ (1 | id),
data = hsball,
family = binomial("logit"))
summary(m0_logit)># Generalized linear mixed model fit by maximum likelihood (Laplace
># Approximation) [glmerMod]
># Family: binomial ( logit )
># Formula: mathcom ~ (1 | id)
># Data: hsball
>#
># AIC BIC logLik deviance df.resid
># 6485.2 6499.0 -3240.6 6481.2 7183
>#
># Scaled residuals:
># Min 1Q Median 3Q Max
># -1.0164 -0.4902 -0.3881 -0.2634 4.0721
>#
># Random effects:
># Groups Name Variance Std.Dev.
># id (Intercept) 0.5554 0.7453
># Number of obs: 7185, groups: id, 160
>#
># Fixed effects:
># Estimate Std. Error z value Pr(>|z|)
># (Intercept) -1.68945 0.06956 -24.29 <2e-16 ***
># ---
># Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1confint(m0_logit) # confidence intervals># Computing profile confidence intervals ...># 2.5 % 97.5 %
># .sig01 0.6346711 0.8765604
># (Intercept) -1.8312696 -1.5552459# Note: Quadrature in glmer() only works for models
# without random slopes
m0_logit_ghq <- glmer(mathcom ~ (1 | id),
data = hsball,
family = binomial("logit"),
nAGQ = 15)
summary(m0_logit_ghq)># Generalized linear mixed model fit by maximum likelihood (Adaptive
># Gauss-Hermite Quadrature, nAGQ = 15) [glmerMod]
># Family: binomial ( logit )
># Formula: mathcom ~ (1 | id)
># Data: hsball
>#
># AIC BIC logLik deviance df.resid
># 6483.9 6497.6 -3239.9 6479.9 7183
>#
># Scaled residuals:
># Min 1Q Median 3Q Max
># -1.0181 -0.4904 -0.3878 -0.2626 4.0871
>#
># Random effects:
># Groups Name Variance Std.Dev.
># id (Intercept) 0.5643 0.7512
># Number of obs: 7185, groups: id, 160
>#
># Fixed effects:
># Estimate Std. Error z value Pr(>|z|)
># (Intercept) -1.69060 0.07034 -24.04 <2e-16 ***
># ---
># Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1confint(m0_logit_ghq) # confidence intervals># Computing profile confidence intervals ...># 2.5 % 97.5 %
># .sig01 0.6392388 0.8835765
># (Intercept) -1.8327418 -1.5550678m0_logit_brm <- brm(mathcom ~ (1 | id),
data = hsball,
family = bernoulli("logit"),
file = "brms_cached_m0_logit")
summary(m0_logit_brm) # include interval estimates># Family: bernoulli
># Links: mu = logit
># Formula: mathcom ~ (1 | id)
># Data: hsball (Number of observations: 7185)
># Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
># total post-warmup draws = 4000
>#
># Group-Level Effects:
># ~id (Number of levels: 160)
># Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
># sd(Intercept) 0.76 0.06 0.65 0.89 1.00 1390 2502
>#
># Population-Level Effects:
># Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
># Intercept -1.69 0.07 -1.83 -1.55 1.00 1728 2337
>#
># Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
># and Tail_ESS are effective sample size measures, and Rhat is the potential
># scale reduction factor on split chains (at convergence, Rhat = 1).By definition,
vc_m0 <- VarCorr(m0_logit)
(icc_m0 <- vc_m0$id[1, 1] /
(vc_m0$id[1, 1] + pi^2 / 3))># [1] 0.1444433# Same with performance::icc()
performance::icc(m0_logit)># # Intraclass Correlation Coefficient
>#
># Adjusted ICC: 0.144
># Unadjusted ICC: 0.144performance::icc(m0_logit_brm)># # Intraclass Correlation Coefficient
>#
># Adjusted ICC: 0.150
># Unadjusted ICC: 0.150With Lv-2 and Lv-1 Predictors
hsball <- hsball %>%
group_by(id) %>% # operate within schools
mutate(ses_cm = mean(ses), # create cluster means (the same as `meanses`)
ses_cmc = ses - ses_cm) %>% # cluster-mean centered
ungroup() # exit the "editing within groups" modeLv-1:
Lv-2:
m1_logit <- glmer(
mathcom ~ meanses + ses_cmc + (ses_cmc | id),
data = hsball,
family = binomial("logit")
)># boundary (singular) fit: see help('isSingular')# There is a singular convergence warning, but the likelihood values
# seem fine with different optimizers.
summary(m1_logit)># Generalized linear mixed model fit by maximum likelihood (Laplace
># Approximation) [glmerMod]
># Family: binomial ( logit )
># Formula: mathcom ~ meanses + ses_cmc + (ses_cmc | id)
># Data: hsball
>#
># AIC BIC logLik deviance df.resid
># 6268.7 6310.0 -3128.3 6256.7 7179
>#
># Scaled residuals:
># Min 1Q Median 3Q Max
># -1.1590 -0.4955 -0.3622 -0.2249 5.6535
>#
># Random effects:
># Groups Name Variance Std.Dev. Corr
># id (Intercept) 0.26670 0.5164
># ses_cmc 0.01275 0.1129 -1.00
># Number of obs: 7185, groups: id, 160
>#
># Fixed effects:
># Estimate Std. Error z value Pr(>|z|)
># (Intercept) -1.75812 0.05659 -31.07 <2e-16 ***
># meanses 1.43528 0.13477 10.65 <2e-16 ***
># ses_cmc 0.60632 0.05553 10.92 <2e-16 ***
># ---
># Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
>#
># Correlation of Fixed Effects:
># (Intr) meanss
># meanses -0.169
># ses_cmc -0.316 0.004
># optimizer (Nelder_Mead) convergence code: 0 (OK)
># boundary (singular) fit: see help('isSingular')Bayesian estimation
One challenge with lme4 is that the estimation may not be accurate for generalized linear mixed-effect models (GLMMs), especially in the presence of random slopes. We saw convergence issues in our previous model. A better approach is to use Bayesian estimation. A good practice is to compare the results from lme4::glmer() to those with Bayesian analysis. If there is a large discrepancy, the Bayesian result is likely more accurate, especially in small samples.
brms::brm()
m1_logit_brm <- brm(
mathcom ~ meanses + ses_cmc + (ses_cmc | id),
data = hsball,
family = bernoulli("logit"),
file = "brms_cached_m1_logit",
# Make results reproducible for Bayesian
seed = 1541)
summary(m1_logit_brm) # include interval estimates># Family: bernoulli
># Links: mu = logit
># Formula: mathcom ~ meanses + ses_cmc + (ses_cmc | id)
># Data: hsball (Number of observations: 7185)
># Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
># total post-warmup draws = 4000
>#
># Group-Level Effects:
># ~id (Number of levels: 160)
># Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS
># sd(Intercept) 0.53 0.05 0.43 0.64 1.00 1444
># sd(ses_cmc) 0.11 0.07 0.01 0.26 1.00 1368
># cor(Intercept,ses_cmc) -0.48 0.44 -0.98 0.67 1.00 2734
># Tail_ESS
># sd(Intercept) 2022
># sd(ses_cmc) 1512
># cor(Intercept,ses_cmc) 2038
>#
># Population-Level Effects:
># Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
># Intercept -1.76 0.06 -1.87 -1.65 1.00 1882 2898
># meanses 1.45 0.14 1.19 1.73 1.00 1820 2529
># ses_cmc 0.59 0.06 0.49 0.70 1.00 4497 2875
>#
># Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
># and Tail_ESS are effective sample size measures, and Rhat is the potential
># scale reduction factor on split chains (at convergence, Rhat = 1).Plotting
Plotting is especially important with transformation, as it shows the association in the probability unit.
m1_plots <- plot_model(m1_logit,
type = "pred",
show.data = TRUE, jitter = 0.02,
title = "", dot.size = 0.5
)># Data were 'prettified'. Consider using `terms="meanses [all]"` to get
># smooth plots.># Data were 'prettified'. Consider using `terms="ses_cmc [all]"` to get
># smooth plots.gridExtra::grid.arrange(grobs = m1_plots, ncol = 2)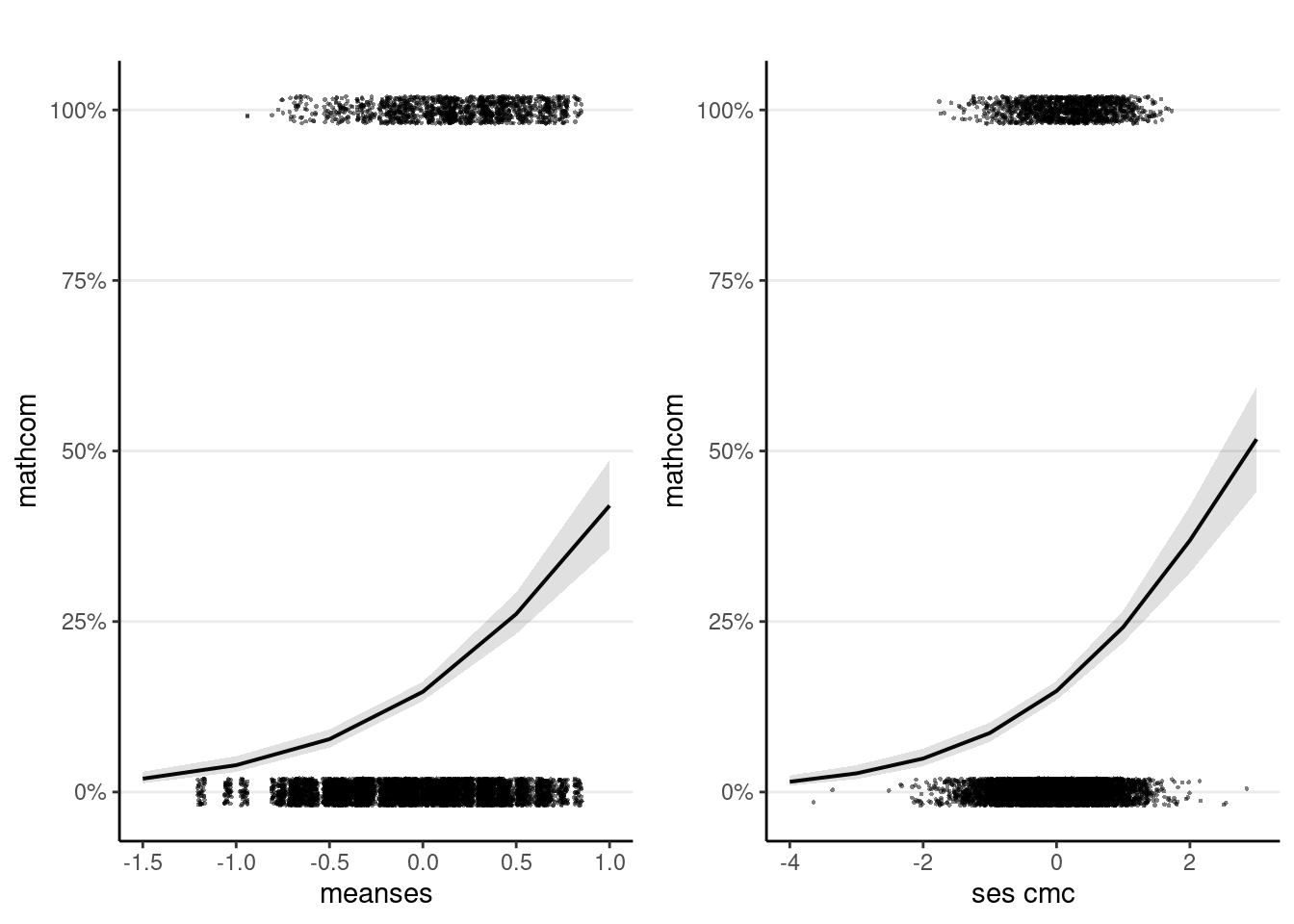
Interpretations
Intercept
For a student with ses = 0 in a school with meanses = 0, the predicted log odds of being commended was -1.76, 95% CI [-1.87, -1.65].
Slopes
A unit increase in meanses is associated with an increase in log odds of 1.44, 95% CI [1.17, 1.71].
Within a given school, a unit increase in student-level ses is associated with an increase in log odds of 0.606, 95% CI [0.498, 0.719].
Odds ratio
A unit increase in meanses is associated with the odds of being commended multiplied by 1.44, 95% CI [3.23, 5.51].
Within a given school, a unit increase in ses is associated with the odds of being commended multiplied by 0.606, 95% CI [1.65, 2.05].
Predicted probabilities with representative values
# meanses = 0; ses_cmc = -0.5 vs 0.5
pred_df1 <- expand_grid(meanses = 0,
ses_cmc = c(-0.5, 0.5),
id = NA)
cbind(pred_df1,
.pred = predict(m1_logit, newdata = pred_df1,
re.form = NA, type = "response"))meanses <dbl> | ses_cmc <dbl> | id <lgl> | .pred <dbl> | |
|---|---|---|---|---|
| 1 | 0 | -0.5 | NA | 0.1129170 |
| 2 | 0 | 0.5 | NA | 0.1892387 |
# meanses = -0.5 vs 0.5; ses_cmc = 0
pred_df2 <- expand_grid(meanses = c(-0.5, 0.5),
ses_cmc = 0,
id = NA)
cbind(pred_df2,
.pred = predict(m1_logit, newdata = pred_df2,
re.form = NA, type = "response"))meanses <dbl> | ses_cmc <dbl> | id <lgl> | .pred <dbl> | |
|---|---|---|---|---|
| 1 | -0.5 | 0 | NA | 0.07757459 |
| 2 | 0.5 | 0 | NA | 0.26105681 |
As shown above, a difference of 1 unit in ses_cmc from -0.5 to 0.5 corresponded to a difference in the probability of being commended by about 7%, when meanses = 0.
On the other hand, a difference of 1 unit in meanses from -0.5 to 0.5 corresponded to a difference in the probability of being commended by about 18%, when ses_cmc = 0.
r.squaredGLMM(m1_logit)># Warning: 'r.squaredGLMM' now calculates a revised statistic. See the help page.># Warning: the null model is correct only if all variables used by the original
># model remain unchanged.># R2m R2c
># theoretical 0.1258308 0.19264569
># delta 0.0628915 0.09628626There are two versions of R^2 being reported. The “theoretical” one refers to the R^2 by assuming that the binary outcome originally comes from a continuous underlying distribution, and so may be appropriate here. The “delta” one looks at the variance of the binary outcome directly, so it is appropriate for outcomes that can only take two values. In practice, either one can be used. See https://royalsocietypublishing.org/doi/pdf/10.1098/rsif.2017.0213 and https://stats.idre.ucla.edu/other/mult-pkg/faq/general/faq-what-are-pseudo-r-squareds/ for more discussions.
Table of Coefficients
msummary(
list("M0" = m0_logit,
"M1 (Laplace)" = m1_logit,
"M1 (MCMC)" = m1_logit_brm),
statistic = "[{conf.low}, {conf.high}]",
shape = effect + term ~ model,
# Make names for lme4 and brms consistent
coef_rename = c("b_Intercept" = "(Intercept)",
"b_meanses" = "meanses",
"b_ses_cmc" = "ses_cmc",
"sd_id__Intercept" = "SD (Intercept id)",
"sd_id__ses_cmc" = "SD (ses_cmc id)",
"cor_id__Intercept__ses_cmc" = "Cor (Intercept~ses_cmc id)"),
gof_map = c("nobs"),
metrics = "none"
)| M0 | M1 (Laplace) | M1 (MCMC) | ||
|---|---|---|---|---|
| fixed | (Intercept) | −1.689 | −1.758 | −1.756 |
| [−1.826, −1.553] | [−1.869, −1.647] | [−1.868, −1.649] | ||
| meanses | 1.435 | 1.449 | ||
| [1.171, 1.699] | [1.187, 1.730] | |||
| ses_cmc | 0.606 | 0.592 | ||
| [0.497, 0.715] | [0.486, 0.702] | |||
| random | SD (Intercept id) | 0.745 | 0.516 | 0.525 |
| [0.428, 0.636] | ||||
| SD (ses_cmc id) | 0.113 | 0.103 | ||
| [0.005, 0.256] | ||||
| Cor (Intercept~ses_cmc id) | −1.000 | −0.602 | ||
| [−0.984, 0.666] | ||||
| Num.Obs. | 7185 | 7185 | 7185 |
Bonus: Cluster-Specific vs. Population-Average Coefficients
In MLM, we need to distinguish two types of interpretations of coefficients:
- Cluster-specific (CS; also called conditional model): For a given cluster, a unit difference in
- Population-average (PA; also called marginal model): At the population level, a unit difference in
In a linear model, the coefficients under CS and under PA are the same, so we did not pay too much attention to the distinction. However, in generalized linear models, such as logistic models, with the same data, one gets different coefficients by fitting a conditional (e.g., MLM) vs. a marginal model (e.g., GEE). I have some discussions at https://quantscience.rbind.io/2020/12/28/unit-specific-vs-population-average-models/. You may also check out an example in this paper: https://journals.sagepub.com/doi/full/10.3102/10769986211017480.
The GLMMadaptive package computes coefficients in both CS and PA. Below is an example showing the difference:
library(GLMMadaptive)>#
># Attaching package: 'GLMMadaptive'># The following object is masked from 'package:lme4':
>#
># negative.binomialm1_pa <- mixed_model(mathcom ~ meanses + ses_cmc,
random = ~ ses_cmc | id,
data = hsball,
family = binomial("logit"))
# Unit-specific
summary(m1_pa)>#
># Call:
># mixed_model(fixed = mathcom ~ meanses + ses_cmc, random = ~ses_cmc |
># id, data = hsball, family = binomial("logit"))
>#
># Data Descriptives:
># Number of Observations: 7185
># Number of Groups: 160
>#
># Model:
># family: binomial
># link: logit
>#
># Fit statistics:
># log.Lik AIC BIC
># -3128.23 6268.46 6286.911
>#
># Random effects covariance matrix:
># StdDev Corr
># (Intercept) 0.5180
># ses_cmc 0.1174 -0.9369
>#
># Fixed effects:
># Estimate Std.Err z-value p-value
># (Intercept) -1.7584 0.0570 -30.8705 < 1e-04
># meanses 1.4370 0.1357 10.5920 < 1e-04
># ses_cmc 0.6057 0.0560 10.8093 < 1e-04
>#
># Integration:
># method: adaptive Gauss-Hermite quadrature rule
># quadrature points: 11
>#
># Optimization:
># method: hybrid EM and quasi-Newton
># converged: TRUE# Population-average
marginal_coefs(m1_pa, std_errors = TRUE, cores = 2)># Estimate Std.Err z-value p-value
># (Intercept) -1.6713 0.0837 -19.9686 < 1e-04
># meanses 1.3913 0.1316 10.5742 < 1e-04
># ses_cmc 0.5497 0.0660 8.3278 < 1e-04The main thing to note is that when interpreting coefficients in MLM using something other than the identity link, one should note the coefficients are conditional on the random effects. Suppose the interest is in making statements about population-average predictions, such as the difference in probabilities of being commended for students with high and low SES, across all schools. In that case, one needs to obtain the population-average coefficients.