library(here) # makes reading data more consistent
library(tidyverse) # for data manipulation and plotting
library(haven) # for importing SPSS/SAS/Stata data
library(simsem) # for adding missing data
library(mice) # for imputation, and checking missing data pattern
library(mitml) # for multilevel imputations
library(lme4) # for multilevel logistic models
library(brms) # for Bayesian multilevel models
library(modelsummary) # for making tablesWeek 13 (R): Imputation
Load Packages and Import Data
Import
The data is the first wave of the Cognition, Health, and Aging Project.
# Download the data from
# https://www.pilesofvariance.com/Chapter8/SPSS/SPSS_Chapter8.zip, and put it
# into the "data_files" folder
zip_data <- here("data_files", "SPSS_Chapter8.zip")
if (!file.exists(zip_data)) {
download.file(
"https://www.pilesofvariance.com/Chapter8/SPSS/SPSS_Chapter8.zip",
zip_data
)
}
stress_data <- read_sav(
unz(zip_data,
"SPSS_Chapter8/SPSS_Chapter8.sav"))
stress_dataPersonID <dbl> | women <dbl> | baseage <dbl> | session <dbl> | studyday <dbl> | dayofweek <dbl> | weekend <dbl> | |
|---|---|---|---|---|---|---|---|
| 102 | 1 | 83.35387 | 2 | 1 | 6 | 1 | |
| 102 | 1 | 83.35387 | 3 | 5 | 3 | 0 | |
| 102 | 1 | 83.35387 | 4 | 7 | 5 | 0 | |
| 102 | 1 | 83.35387 | 5 | 8 | 6 | 1 | |
| 102 | 1 | 83.35387 | 6 | 9 | 7 | 1 | |
| 103 | 1 | 92.00000 | 2 | 1 | 6 | 1 | |
| 103 | 1 | 92.00000 | 3 | 2 | 7 | 1 | |
| 103 | 1 | 92.00000 | 4 | 3 | 1 | 0 | |
| 103 | 1 | 92.00000 | 5 | 4 | 2 | 0 | |
| 103 | 1 | 92.00000 | 6 | 8 | 6 | 1 |
Analysis With Original Data
# Compute cluster means
stress_data <- stress_data %>%
# Center mood (originally 1-5) at 1 for interpretation (so it becomes 0-4)
# Also recode women, weekend, and stressor to factor
mutate(mood1 = mood - 1,
women = factor(women, levels = c(0, 1),
labels = c("men", "women"))) %>%
group_by(PersonID) %>%
# The `across()` function applies the same procedure on
# multiple variables
mutate(across(c(symptoms, mood1, stressor),
# The `.` means the variable to be operated on
list("pm" = ~ mean(., na.rm = TRUE),
"pmc" = ~ . - mean(., na.rm = TRUE)))) %>%
ungroup()
m1 <- lmer(
symptoms ~ mood1_pm + mood1_pmc + women + (mood1_pmc | PersonID),
data = stress_data
)># boundary (singular) fit: see help('isSingular')summary(m1)># Linear mixed model fit by REML ['lmerMod']
># Formula: symptoms ~ mood1_pm + mood1_pmc + women + (mood1_pmc | PersonID)
># Data: stress_data
>#
># REML criterion at convergence: 1430.5
>#
># Scaled residuals:
># Min 1Q Median 3Q Max
># -3.1983 -0.4930 -0.1134 0.4272 4.0405
>#
># Random effects:
># Groups Name Variance Std.Dev. Corr
># PersonID (Intercept) 0.88812 0.9424
># mood1_pmc 0.03862 0.1965 1.00
># Residual 0.61483 0.7841
># Number of obs: 514, groups: PersonID, 105
>#
># Fixed effects:
># Estimate Std. Error t value
># (Intercept) 1.2263 0.2024 6.060
># mood1_pm 2.1231 0.3641 5.831
># mood1_pmc 0.1330 0.1295 1.027
># womenwomen -0.4955 0.2207 -2.245
>#
># Correlation of Fixed Effects:
># (Intr) mod1_pm md1_pmc
># mood1_pm -0.350
># mood1_pmc -0.005 0.179
># womenwomen -0.792 -0.025 0.007
># optimizer (nloptwrap) convergence code: 0 (OK)
># boundary (singular) fit: see help('isSingular')Missing Data Handling
Add 20% missing data based on MAR
set.seed(1525)
stress_data_mis <- simsem::imposeMissing(
stress_data[1:10],
cov = "baseage",
pmMAR = .2,
ignoreCols = 1:7
)mice::md.pattern(stress_data_mis[1:10], rotate.names = TRUE)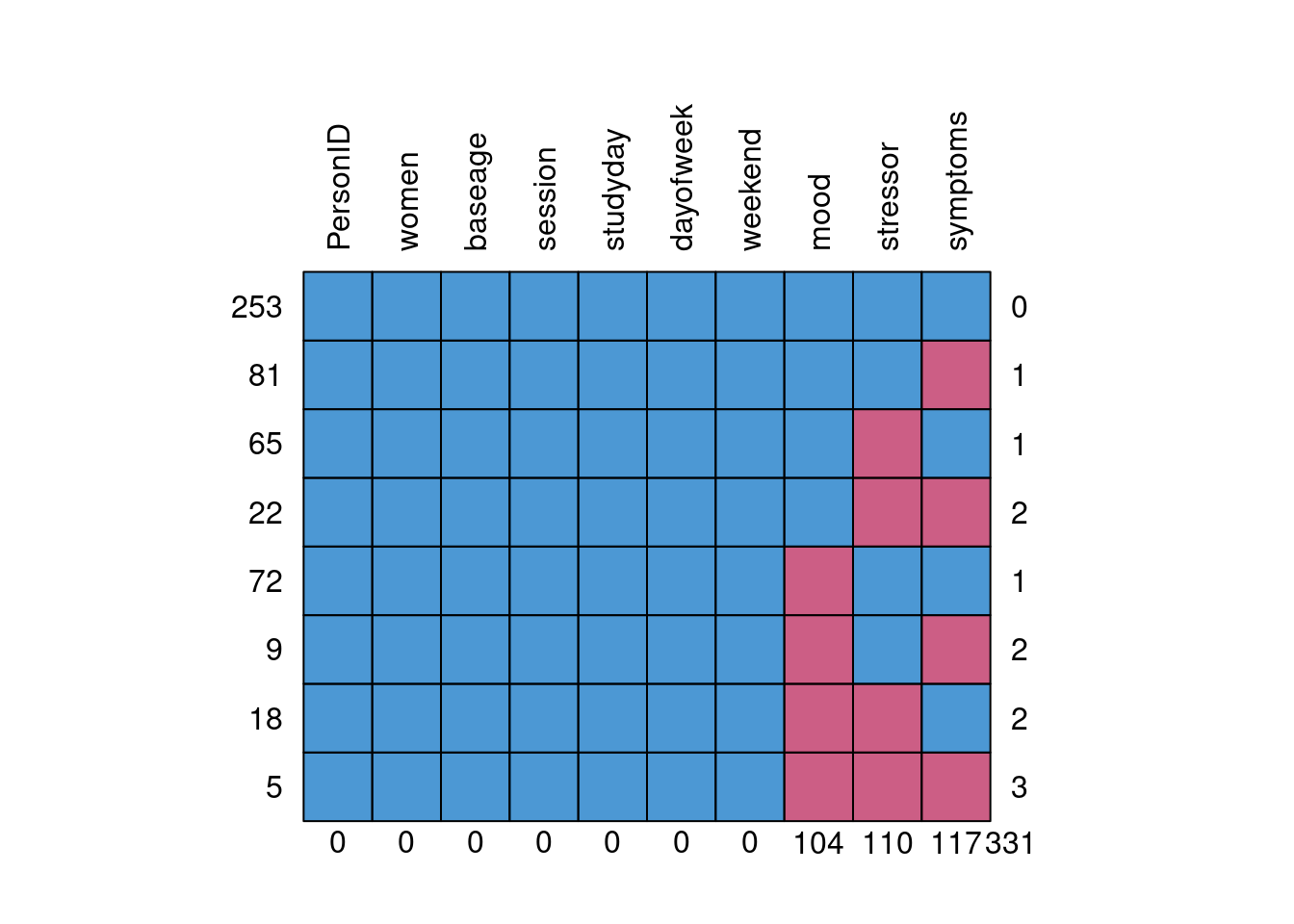
># PersonID women baseage session studyday dayofweek weekend mood stressor
># 253 1 1 1 1 1 1 1 1 1
># 81 1 1 1 1 1 1 1 1 1
># 65 1 1 1 1 1 1 1 1 0
># 22 1 1 1 1 1 1 1 1 0
># 72 1 1 1 1 1 1 1 0 1
># 9 1 1 1 1 1 1 1 0 1
># 18 1 1 1 1 1 1 1 0 0
># 5 1 1 1 1 1 1 1 0 0
># 0 0 0 0 0 0 0 104 110
># symptoms
># 253 1 0
># 81 0 1
># 65 1 1
># 22 0 2
># 72 1 1
># 9 0 2
># 18 1 2
># 5 0 3
># 117 331Listwise Deletion
# Compute cluster means
stress_data_mis <- stress_data_mis %>%
# Center mood (originally 1-5) at 1 for interpretation (so it becomes 0-4)
# Also recode women to factor
mutate(mood1 = mood - 1) %>%
group_by(PersonID) %>%
# The `across()` function applies the same procedure on
# multiple variables
mutate(across(c(symptoms, mood1, stressor),
# The `.` means the variable to be operated on
list("pm" = ~ mean(., na.rm = TRUE),
"pmc" = ~ . - mean(., na.rm = TRUE)))) %>%
ungroup()
m1_lw <- lmer(
symptoms ~ mood1_pm + mood1_pmc + women + (mood1_pmc | PersonID),
data = stress_data_mis
)># boundary (singular) fit: see help('isSingular')summary(m1_lw)># Linear mixed model fit by REML ['lmerMod']
># Formula: symptoms ~ mood1_pm + mood1_pmc + women + (mood1_pmc | PersonID)
># Data: stress_data_mis
>#
># REML criterion at convergence: 891.8
>#
># Scaled residuals:
># Min 1Q Median 3Q Max
># -3.3129 -0.4996 -0.1467 0.4517 2.8819
>#
># Random effects:
># Groups Name Variance Std.Dev. Corr
># PersonID (Intercept) 0.84314 0.9182
># mood1_pmc 0.08967 0.2994 1.00
># Residual 0.55069 0.7421
># Number of obs: 318, groups: PersonID, 105
>#
># Fixed effects:
># Estimate Std. Error t value
># (Intercept) 1.2813 0.2005 6.391
># mood1_pm 2.0700 0.3456 5.990
># mood1_pmc 0.2856 0.1648 1.733
># womenwomen -0.6635 0.2224 -2.984
>#
># Correlation of Fixed Effects:
># (Intr) mod1_pm md1_pmc
># mood1_pm -0.321
># mood1_pmc -0.019 0.275
># womenwomen -0.779 -0.073 -0.001
># optimizer (nloptwrap) convergence code: 0 (OK)
># boundary (singular) fit: see help('isSingular')Multiple Imputation
Need to recode categorical variables to factor first
to_impute <- stress_data_mis[1:10] %>%
mutate(weekend = factor(weekend, levels = c(0, 1),
labels = c("no", "yes")),
stressor = factor(stressor, levels = c(0, 1),
labels = c("no", "yes")))
imp <- jomoImpute(
to_impute,
formula = mood + stressor + symptoms ~
women + baseage + weekend + (1 | PersonID),
n.burn = 5000,
n.iter = 100,
m = 100
)># Running burn-in phase ...
># Creating imputed data set ( 1 / 100 ) ...
># Creating imputed data set ( 2 / 100 ) ...
># Creating imputed data set ( 3 / 100 ) ...
># Creating imputed data set ( 4 / 100 ) ...
># Creating imputed data set ( 5 / 100 ) ...
># Creating imputed data set ( 6 / 100 ) ...
># Creating imputed data set ( 7 / 100 ) ...
># Creating imputed data set ( 8 / 100 ) ...
># Creating imputed data set ( 9 / 100 ) ...
># Creating imputed data set ( 10 / 100 ) ...
># Creating imputed data set ( 11 / 100 ) ...
># Creating imputed data set ( 12 / 100 ) ...
># Creating imputed data set ( 13 / 100 ) ...
># Creating imputed data set ( 14 / 100 ) ...
># Creating imputed data set ( 15 / 100 ) ...
># Creating imputed data set ( 16 / 100 ) ...
># Creating imputed data set ( 17 / 100 ) ...
># Creating imputed data set ( 18 / 100 ) ...
># Creating imputed data set ( 19 / 100 ) ...
># Creating imputed data set ( 20 / 100 ) ...
># Creating imputed data set ( 21 / 100 ) ...
># Creating imputed data set ( 22 / 100 ) ...
># Creating imputed data set ( 23 / 100 ) ...
># Creating imputed data set ( 24 / 100 ) ...
># Creating imputed data set ( 25 / 100 ) ...
># Creating imputed data set ( 26 / 100 ) ...
># Creating imputed data set ( 27 / 100 ) ...
># Creating imputed data set ( 28 / 100 ) ...
># Creating imputed data set ( 29 / 100 ) ...
># Creating imputed data set ( 30 / 100 ) ...
># Creating imputed data set ( 31 / 100 ) ...
># Creating imputed data set ( 32 / 100 ) ...
># Creating imputed data set ( 33 / 100 ) ...
># Creating imputed data set ( 34 / 100 ) ...
># Creating imputed data set ( 35 / 100 ) ...
># Creating imputed data set ( 36 / 100 ) ...
># Creating imputed data set ( 37 / 100 ) ...
># Creating imputed data set ( 38 / 100 ) ...
># Creating imputed data set ( 39 / 100 ) ...
># Creating imputed data set ( 40 / 100 ) ...
># Creating imputed data set ( 41 / 100 ) ...
># Creating imputed data set ( 42 / 100 ) ...
># Creating imputed data set ( 43 / 100 ) ...
># Creating imputed data set ( 44 / 100 ) ...
># Creating imputed data set ( 45 / 100 ) ...
># Creating imputed data set ( 46 / 100 ) ...
># Creating imputed data set ( 47 / 100 ) ...
># Creating imputed data set ( 48 / 100 ) ...
># Creating imputed data set ( 49 / 100 ) ...
># Creating imputed data set ( 50 / 100 ) ...
># Creating imputed data set ( 51 / 100 ) ...
># Creating imputed data set ( 52 / 100 ) ...
># Creating imputed data set ( 53 / 100 ) ...
># Creating imputed data set ( 54 / 100 ) ...
># Creating imputed data set ( 55 / 100 ) ...
># Creating imputed data set ( 56 / 100 ) ...
># Creating imputed data set ( 57 / 100 ) ...
># Creating imputed data set ( 58 / 100 ) ...
># Creating imputed data set ( 59 / 100 ) ...
># Creating imputed data set ( 60 / 100 ) ...
># Creating imputed data set ( 61 / 100 ) ...
># Creating imputed data set ( 62 / 100 ) ...
># Creating imputed data set ( 63 / 100 ) ...
># Creating imputed data set ( 64 / 100 ) ...
># Creating imputed data set ( 65 / 100 ) ...
># Creating imputed data set ( 66 / 100 ) ...
># Creating imputed data set ( 67 / 100 ) ...
># Creating imputed data set ( 68 / 100 ) ...
># Creating imputed data set ( 69 / 100 ) ...
># Creating imputed data set ( 70 / 100 ) ...
># Creating imputed data set ( 71 / 100 ) ...
># Creating imputed data set ( 72 / 100 ) ...
># Creating imputed data set ( 73 / 100 ) ...
># Creating imputed data set ( 74 / 100 ) ...
># Creating imputed data set ( 75 / 100 ) ...
># Creating imputed data set ( 76 / 100 ) ...
># Creating imputed data set ( 77 / 100 ) ...
># Creating imputed data set ( 78 / 100 ) ...
># Creating imputed data set ( 79 / 100 ) ...
># Creating imputed data set ( 80 / 100 ) ...
># Creating imputed data set ( 81 / 100 ) ...
># Creating imputed data set ( 82 / 100 ) ...
># Creating imputed data set ( 83 / 100 ) ...
># Creating imputed data set ( 84 / 100 ) ...
># Creating imputed data set ( 85 / 100 ) ...
># Creating imputed data set ( 86 / 100 ) ...
># Creating imputed data set ( 87 / 100 ) ...
># Creating imputed data set ( 88 / 100 ) ...
># Creating imputed data set ( 89 / 100 ) ...
># Creating imputed data set ( 90 / 100 ) ...
># Creating imputed data set ( 91 / 100 ) ...
># Creating imputed data set ( 92 / 100 ) ...
># Creating imputed data set ( 93 / 100 ) ...
># Creating imputed data set ( 94 / 100 ) ...
># Creating imputed data set ( 95 / 100 ) ...
># Creating imputed data set ( 96 / 100 ) ...
># Creating imputed data set ( 97 / 100 ) ...
># Creating imputed data set ( 98 / 100 ) ...
># Creating imputed data set ( 99 / 100 ) ...
># Creating imputed data set ( 100 / 100 ) ...
># Done!Assess convergence
plot(imp) # needs good mixingComplete data
implist <- mitmlComplete(imp, "all")
# Centering
implist <- within(implist, {
mood1 <- mood - 1
mood1_pm <- clusterMeans(mood1, cluster = PersonID)
mood1_pmc <- mood1 - mood1_pm
})
m1_mi <- with(
implist,
lmer(symptoms ~ mood1_pm + mood1_pmc + women +
(mood1_pmc | PersonID))
)># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')
># boundary (singular) fit: see help('isSingular')testEstimates(m1_mi, extra.pars = TRUE)>#
># Call:
>#
># testEstimates(model = m1_mi, extra.pars = TRUE)
>#
># Final parameter estimates and inferences obtained from 100 imputed data sets.
>#
># Estimate Std.Error t.value df P(>|t|) RIV FMI
># (Intercept) 1.242 0.203 6.131 41925.356 0.000 0.051 0.049
># mood1_pm 2.024 0.366 5.536 10870.208 0.000 0.106 0.096
># mood1_pmc 0.277 0.159 1.737 629.605 0.083 0.657 0.398
># womenwomen -0.560 0.225 -2.491 27317.284 0.013 0.064 0.060
>#
># Estimate
># Intercept~~Intercept|PersonID 0.873
># mood1_pmc~~mood1_pmc|PersonID 0.067
># Intercept~~mood1_pmc|PersonID 0.158
># Residual~~Residual 0.569
># ICC|PersonID 0.605
>#
># Unadjusted hypothesis test as appropriate in larger samples.