# To install a package, run the following ONCE (and only once on your computer)
# install.packages("psych")
library(here) # makes reading data more consistent
library(tidyverse) # for data manipulation and plotting
library(haven) # for importing SPSS/SAS/Stata data
library(lme4) # for multilevel analysis
library(brms) # for Bayesian multilevel analysis
library(lattice) # for dotplot (working with lme4)
library(sjPlot) # for plotting effects
library(MuMIn) # for computing r-squared
library(r2mlm) # for computing r-squared
library(modelsummary) # for making tablesWeek 3 (R): Random Intercept
Load Packages and Import Data
You can use the message = FALSE option to suppress the package loading messages.
In R, there are many packages for multilevel modeling; two of the most common ones are the lme4 package and the nlme package. In this note, I will show how to run different basic multilevel models using the lme4 package, which is newer. However, some models, like unstructured covariance structure, will need the nlme package or other packages (like the brms package with Bayesian estimation).
Import Data
First, download the data from https://github.com/marklhc/marklai-pages/raw/master/data_files/hsball.sav. We’ll import the data in .sav format using the read_sav() function from the haven package.
# Read in the data (pay attention to the directory)
hsball <- read_sav(here("data_files", "hsball.sav"))
hsball # print the dataid <chr> | minority <dbl> | female <dbl> | ses <dbl> | mathach <dbl> | size <dbl> | sector <dbl> | pracad <dbl> | |
|---|---|---|---|---|---|---|---|---|
| 1224 | 0 | 1 | -1.528 | 5.876 | 842 | 0 | 0.35 | |
| 1224 | 0 | 1 | -0.588 | 19.708 | 842 | 0 | 0.35 | |
| 1224 | 0 | 0 | -0.528 | 20.349 | 842 | 0 | 0.35 | |
| 1224 | 0 | 0 | -0.668 | 8.781 | 842 | 0 | 0.35 | |
| 1224 | 0 | 0 | -0.158 | 17.898 | 842 | 0 | 0.35 | |
| 1224 | 0 | 0 | 0.022 | 4.583 | 842 | 0 | 0.35 | |
| 1224 | 0 | 1 | -0.618 | -2.832 | 842 | 0 | 0.35 | |
| 1224 | 0 | 0 | -0.998 | 0.523 | 842 | 0 | 0.35 | |
| 1224 | 0 | 1 | -0.888 | 1.527 | 842 | 0 | 0.35 | |
| 1224 | 0 | 0 | -0.458 | 21.521 | 842 | 0 | 0.35 |
Run the Random Intercept Model
Model equations
Lv-1:
Lv-2:
Running the model in R
The lme4 package requires input in the format of
outcome ~ fixed + (random | cluster ID)For our data, the combined equation is
- outcome =
mathach, - fixed =
1, - random =
1, and - cluster ID =
id.
Thus the following syntax:
About
brms: Thebrmspackage is a very powerful package that relies on Bayesian estimation, which we will discuss later in the class. I want to start introducing it because, moving forward, there are models that are difficult or impossible to estimate withlme4. If you cannot getbrmsto work, you may need to follow the instruction, which is different depending on your operating system, at https://github.com/stan-dev/rstan/wiki/RStan-Getting-Started to configure C++ Toolchain.
# outcome = mathach
# fixed = gamma_{00} * 1
# random = u_{0j} * 1, with j indexing school id
ran_int <- lmer(mathach ~ 1 + (1 | id), data = hsball)
# Summarize results
summary(ran_int)Linear mixed model fit by REML ['lmerMod']
Formula: mathach ~ 1 + (1 | id)
Data: hsball
REML criterion at convergence: 47116.8
Scaled residuals:
Min 1Q Median 3Q Max
-3.0631 -0.7539 0.0267 0.7606 2.7426
Random effects:
Groups Name Variance Std.Dev.
id (Intercept) 8.614 2.935
Residual 39.148 6.257
Number of obs: 7185, groups: id, 160
Fixed effects:
Estimate Std. Error t value
(Intercept) 12.6370 0.2444 51.71# outcome = mathach
# fixed = gamma_{00} * 1
# random = u_{0j} * 1, with j indexing school id
ran_int_brm <- brm(mathach ~ 1 + (1 | id), data = hsball,
# cache results as brms takes a long time to run
file = "brms_cached_ran_int")
# Summarize results
summary(ran_int_brm) Family: gaussian
Links: mu = identity; sigma = identity
Formula: mathach ~ 1 + (1 | id)
Data: hsball (Number of observations: 7185)
Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
total post-warmup draws = 4000
Group-Level Effects:
~id (Number of levels: 160)
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sd(Intercept) 2.95 0.18 2.62 3.32 1.00 1153 2213
Population-Level Effects:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
Intercept 12.63 0.26 12.11 13.12 1.01 489 805
Family Specific Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sigma 6.26 0.05 6.15 6.36 1.00 8936 2831
Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).Showing the variations across (a subset of) schools
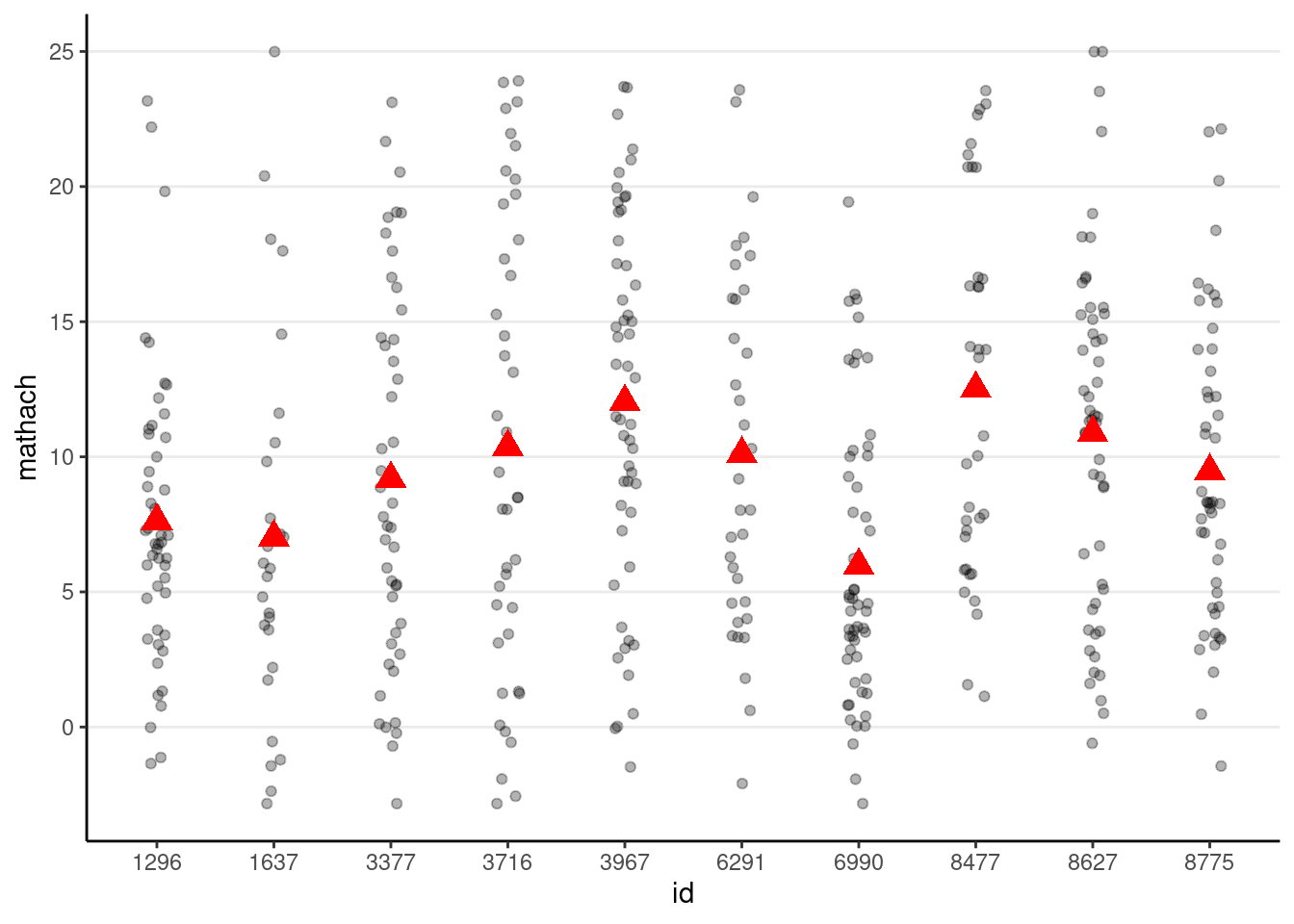
# Randomly select 10 school ids
set.seed(1330) # setting the seed for reproducibility
random_ids <- sample(unique(hsball$id), size = 10)
(p_subset <- hsball %>%
filter(id %in% random_ids) %>% # select only 10 schools
ggplot(aes(x = id, y = mathach)) +
geom_jitter(height = 0, width = 0.1, alpha = 0.3) +
# Add school means
stat_summary(
fun = "mean", geom = "point", col = "red",
shape = 17, # use triangles
size = 4 # make them larger
)
)
Simulating data based on the random intercept model
gamma00 <- 12.6370
tau0 <- 2.935
sigma <- 6.257
num_students <- nrow(hsball)
num_schools <- length(unique(hsball$id))
# Simulate with only gamma00 (i.e., tau0 = 0 and sigma = 0)
simulated_data1 <- tibble(
id = hsball$id,
mathach = gamma00
)
# Show data with no variation
# The `%+%` operator is used to substitute with a different data set
p_subset %+%
(simulated_data1 %>%
filter(id %in% random_ids))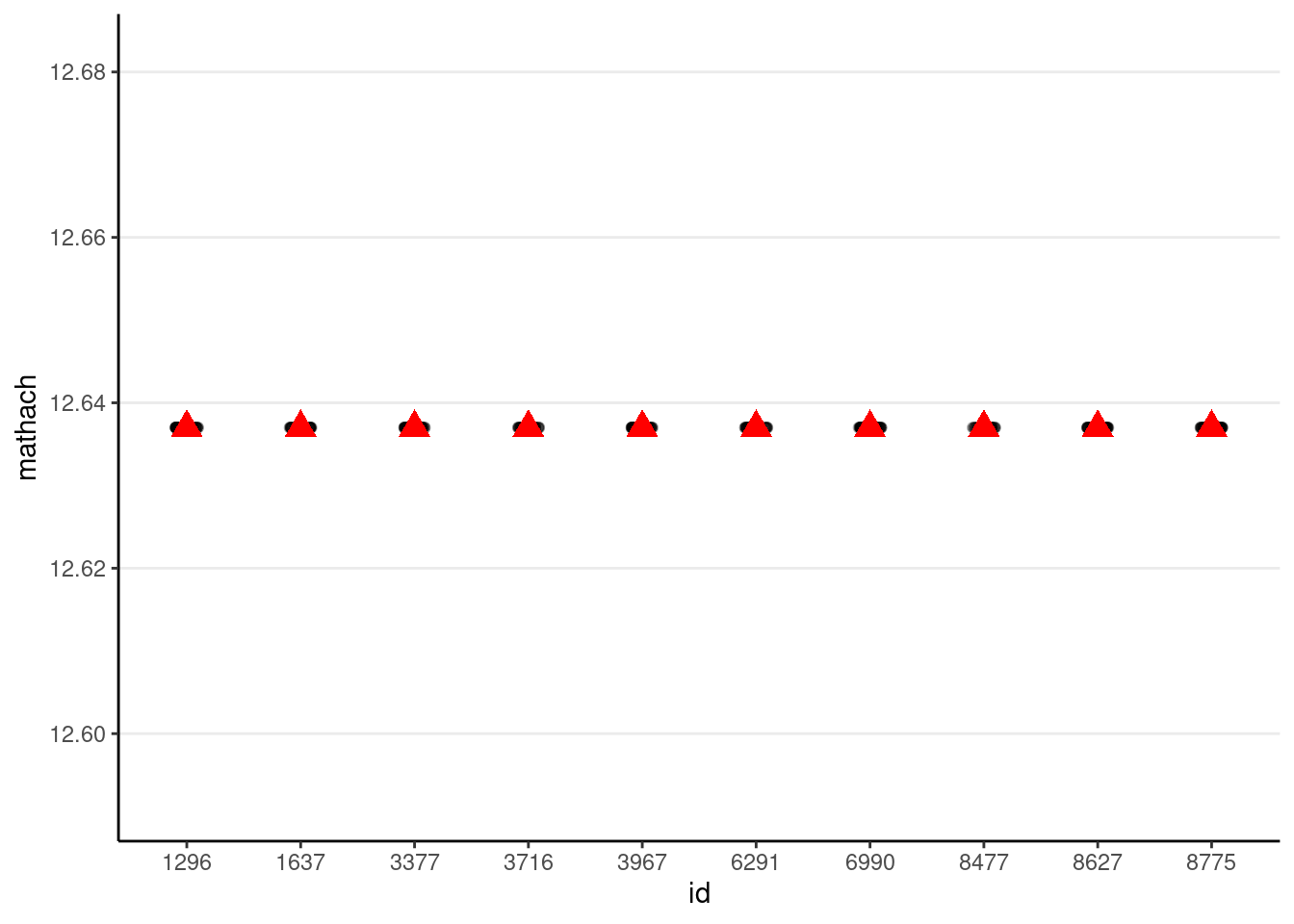
# Simulate with gamma00 + e_ij (i.e., tau0 = 0)
simulated_data2 <- tibble(
id = hsball$id,
mathach = gamma00 + rnorm(num_students, sd = sigma)
)
# Show data with no school-level variation
p_subset %+%
(simulated_data2 %>%
filter(id %in% random_ids))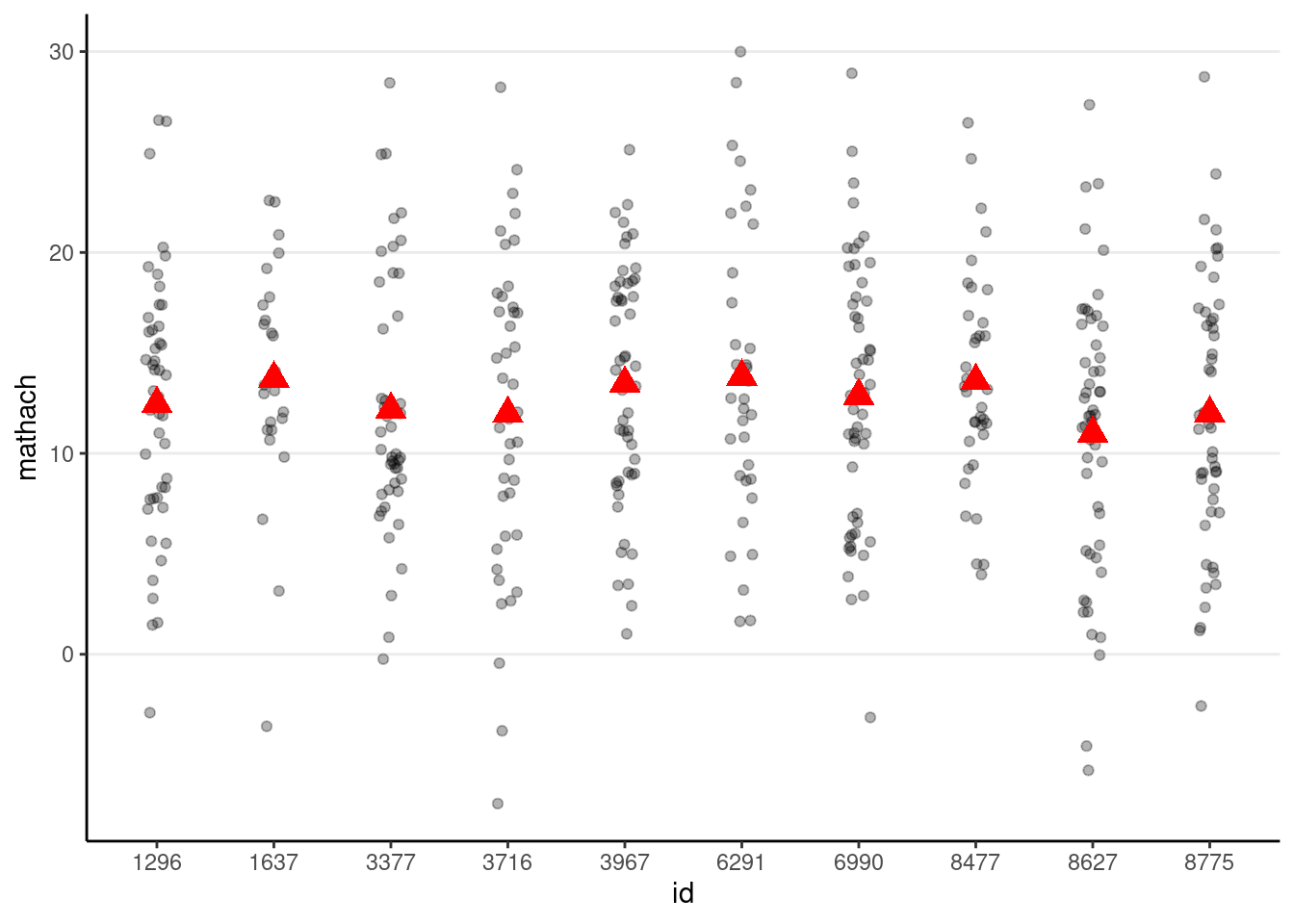
# Simulate with gamma00 + u_0j + e_ij
# First, obtain group indices that start from 1 to 160
group_idx <- group_by(hsball, id) %>% group_indices()
# Then simulate 160 u0j
u0j <- rnorm(num_schools, sd = tau0)
simulated_data3 <- tibble(
id = hsball$id,
mathach = gamma00 +
u0j[group_idx] + # expand the u0j's from 160 to 7185
rnorm(num_students, sd = sigma)
)
# Show data with both school and student variations
p_subset %+%
(simulated_data3 %>%
filter(id %in% random_ids))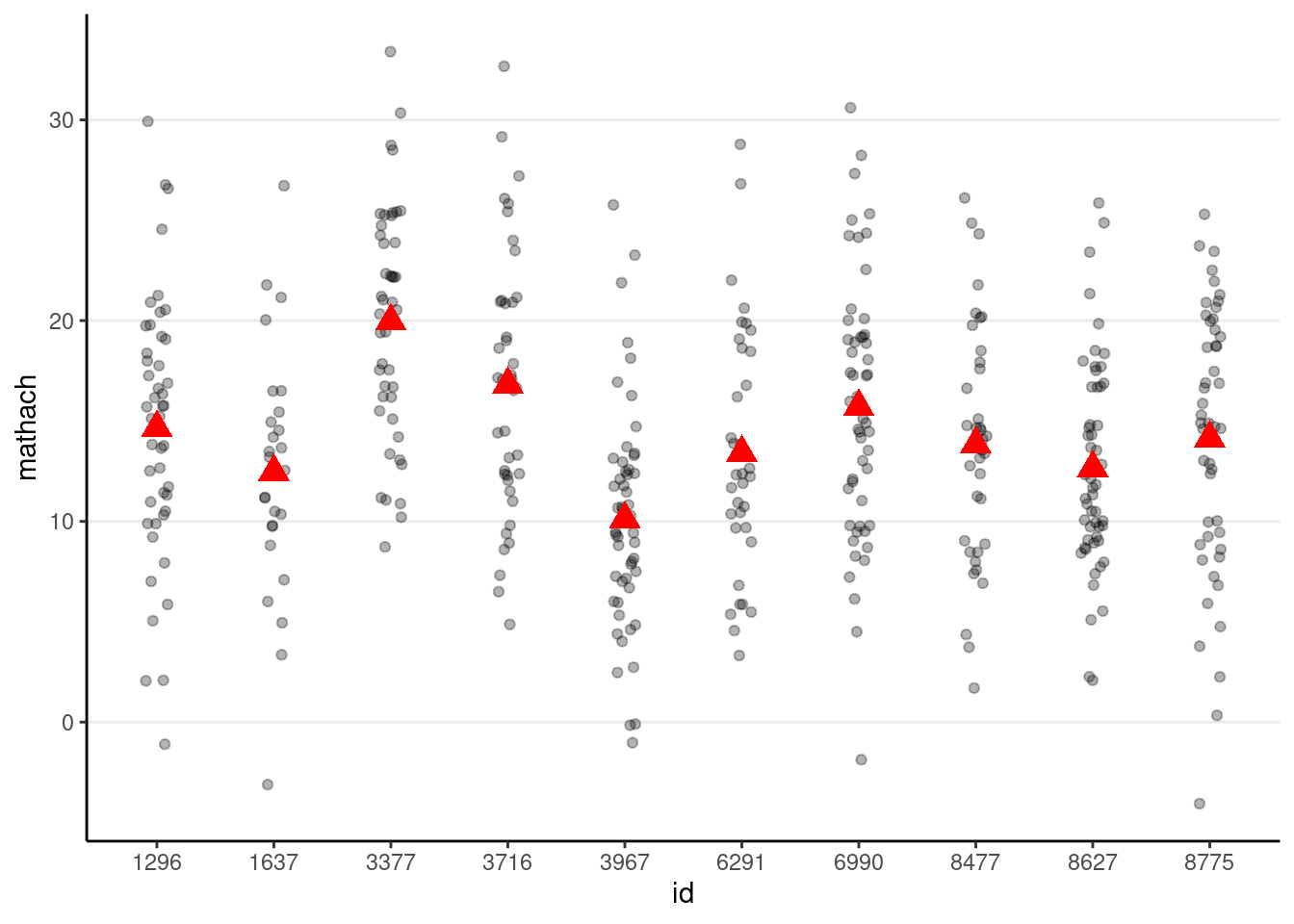
The handy simulate() function can also be used to simulate the data
simulated_math <- simulate(ran_int, nsim = 1)
simulated_data4 <- tibble(
id = hsball$id,
mathach = simulated_math$sim_1
)
p_subset %+%
(simulated_data4 %>%
filter(id %in% random_ids))Plotting the random effects (i.e.,
You can easily plot the estimated school means (also called BLUP, best linear unbiased predictor, or the empirical Bayes (EB) estimates, which are different from the mean of the sample observations for a particular school) using the lattice package:
dotplot(ranef(ran_int, condVar = TRUE))$id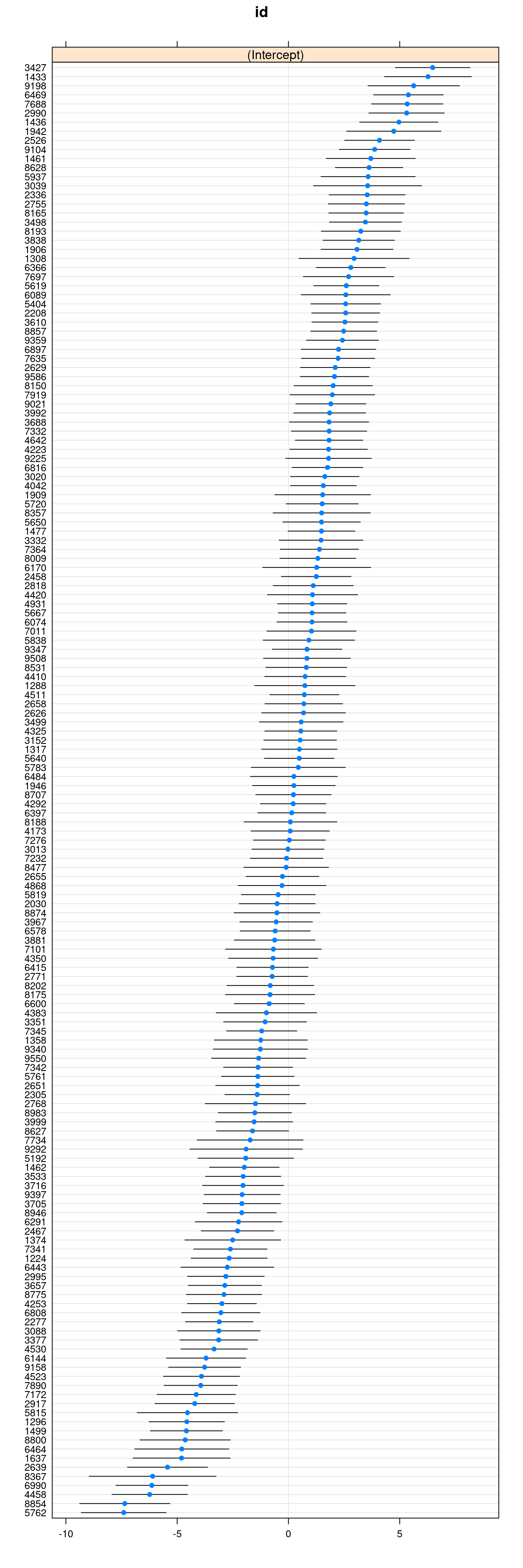
Or with sjPlot::plot_model()
plot_model(ran_int, type = "re")Warning in checkMatrixPackageVersion(): Package version inconsistency detected.
TMB was built with Matrix version 1.5.1
Current Matrix version is 1.5.3
Please re-install 'TMB' from source using install.packages('TMB', type = 'source') or ask CRAN for a binary version of 'TMB' matching CRAN's 'Matrix' package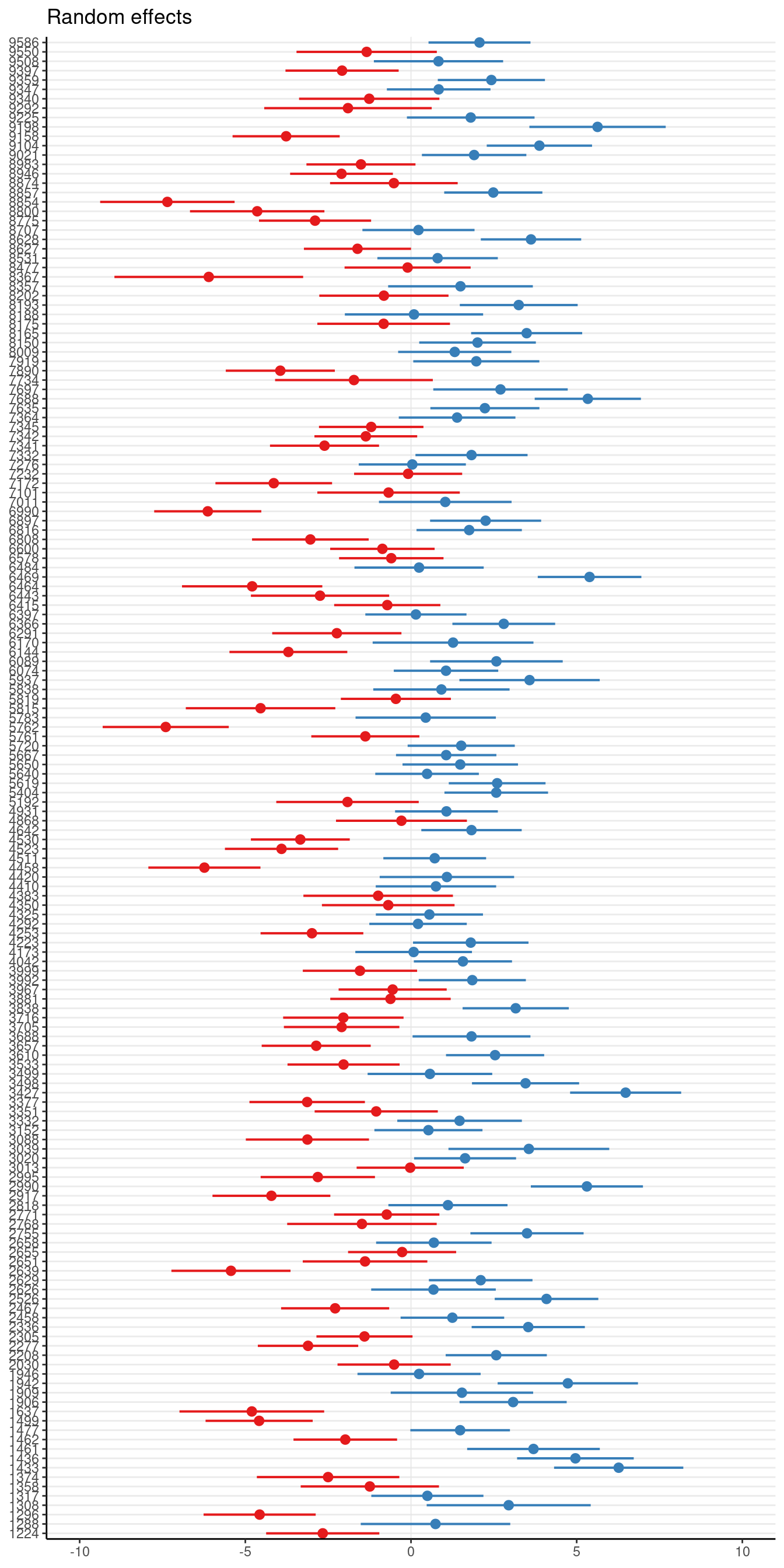
Here’s a plot showing the sample school means (with no borrowing of information) vs. the EB means (borrowing information).
# Compute raw school means and EB means
hsball %>%
group_by(id) %>%
# Raw means
summarise(mathach_raw_means = mean(mathach)) %>%
arrange(mathach_raw_means) %>% # sort by the means
# EB means (the "." means using the current data)
mutate(
mathach_eb_means = predict(ran_int, .),
index = row_number() # add row number as index for plotting
) %>%
ggplot(aes(x = index, y = mathach_raw_means)) +
geom_point(aes(col = "Raw")) +
# Add EB means
geom_point(aes(y = mathach_eb_means, col = "EB"), shape = 1) +
geom_segment(aes(
x = index, xend = index,
y = mathach_eb_means, yend = mathach_raw_means,
col = "EB"
)) +
labs(y = "School mean achievement", col = "")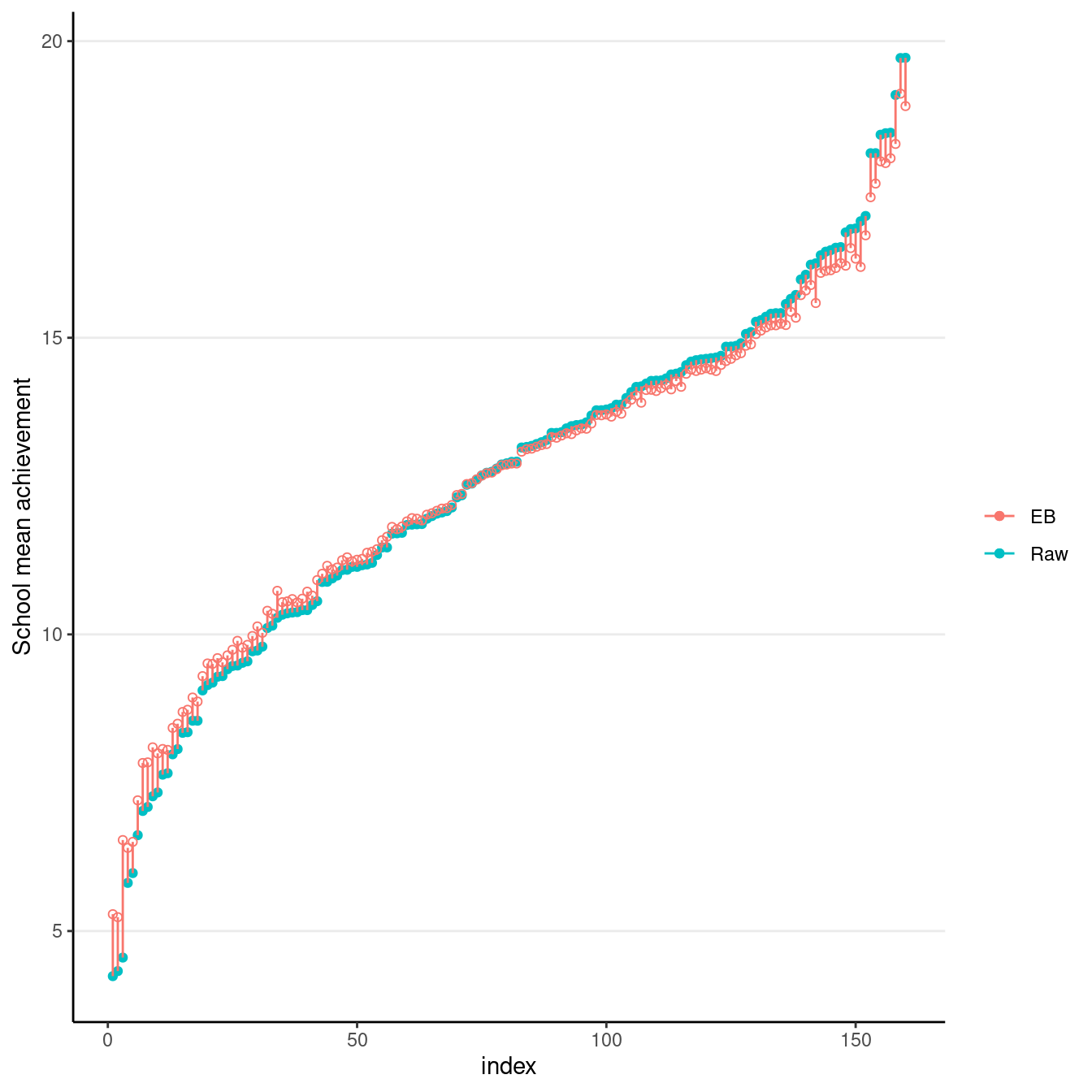
Intraclass correlations
variance_components <- as.data.frame(VarCorr(ran_int))
between_var <- variance_components$vcov[1]
within_var <- variance_components$vcov[2]
(icc <- between_var / (between_var + within_var))[1] 0.1803518# 95% confidence intervals (require installing the bootmlm package)
# if (!require("remotes")) {
# install.packages("remotes")
# }
# remotes::install_github("marklhc/bootmlm")
bootmlm:::prof_ci_icc(ran_int) 2.5 % 97.5 %
0.1471784 0.2210131 Adding Lv-2 Predictors
We have one predictor, meanses, in the fixed part.
hsball %>%
ggplot(aes(x = meanses, y = mathach, col = id)) +
geom_point(alpha = 0.5, size = 0.5) +
guides(col = "none")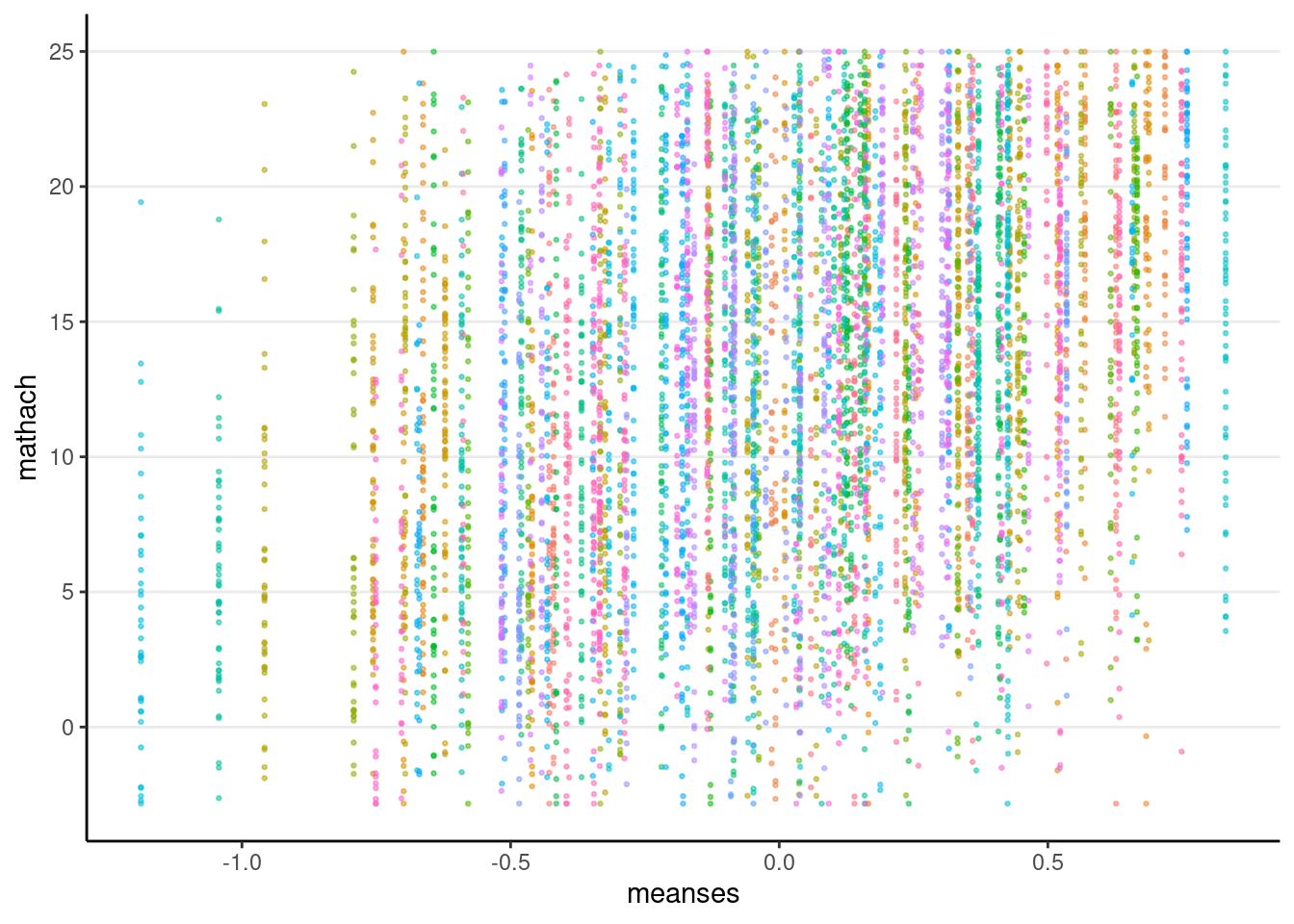
Model Equation
Lv-1:
Lv-2:
meanses that represents the expected difference in school mean achievement between two schools with one unit difference in meanses, and
m_lv2 <- lmer(mathach ~ meanses + (1 | id), data = hsball)
summary(m_lv2)Linear mixed model fit by REML ['lmerMod']
Formula: mathach ~ meanses + (1 | id)
Data: hsball
REML criterion at convergence: 46961.3
Scaled residuals:
Min 1Q Median 3Q Max
-3.13480 -0.75256 0.02409 0.76773 2.78501
Random effects:
Groups Name Variance Std.Dev.
id (Intercept) 2.639 1.624
Residual 39.157 6.258
Number of obs: 7185, groups: id, 160
Fixed effects:
Estimate Std. Error t value
(Intercept) 12.6494 0.1493 84.74
meanses 5.8635 0.3615 16.22
Correlation of Fixed Effects:
(Intr)
meanses -0.004# Likelihood-based confidence intervals for fixed effects
# `parm = "beta_"` requests confidence intervals only for the fixed effects
confint(m_lv2, parm = "beta_")Computing profile confidence intervals ... 2.5 % 97.5 %
(Intercept) 12.356615 12.941707
meanses 5.155769 6.572415m_lv2_brm <- brm(mathach ~ meanses + (1 | id), data = hsball,
# cache results as brms takes a long time to run
file = "brms_cached_m_lv2")
summary(m_lv2_brm) Family: gaussian
Links: mu = identity; sigma = identity
Formula: mathach ~ meanses + (1 | id)
Data: hsball (Number of observations: 7185)
Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
total post-warmup draws = 4000
Group-Level Effects:
~id (Number of levels: 160)
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sd(Intercept) 1.64 0.13 1.40 1.91 1.00 1485 2069
Population-Level Effects:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
Intercept 12.65 0.15 12.37 12.94 1.00 1961 2566
meanses 5.86 0.37 5.16 6.57 1.00 2126 2836
Family Specific Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sigma 6.26 0.05 6.16 6.36 1.00 9510 3281
Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).The 95% confidence intervals (CIs) above showed the uncertainty associated with the estimates. Also, as the 95% CI for meanses does not contain zero, there is evidence for the positive association of SES and mathach at the school level.
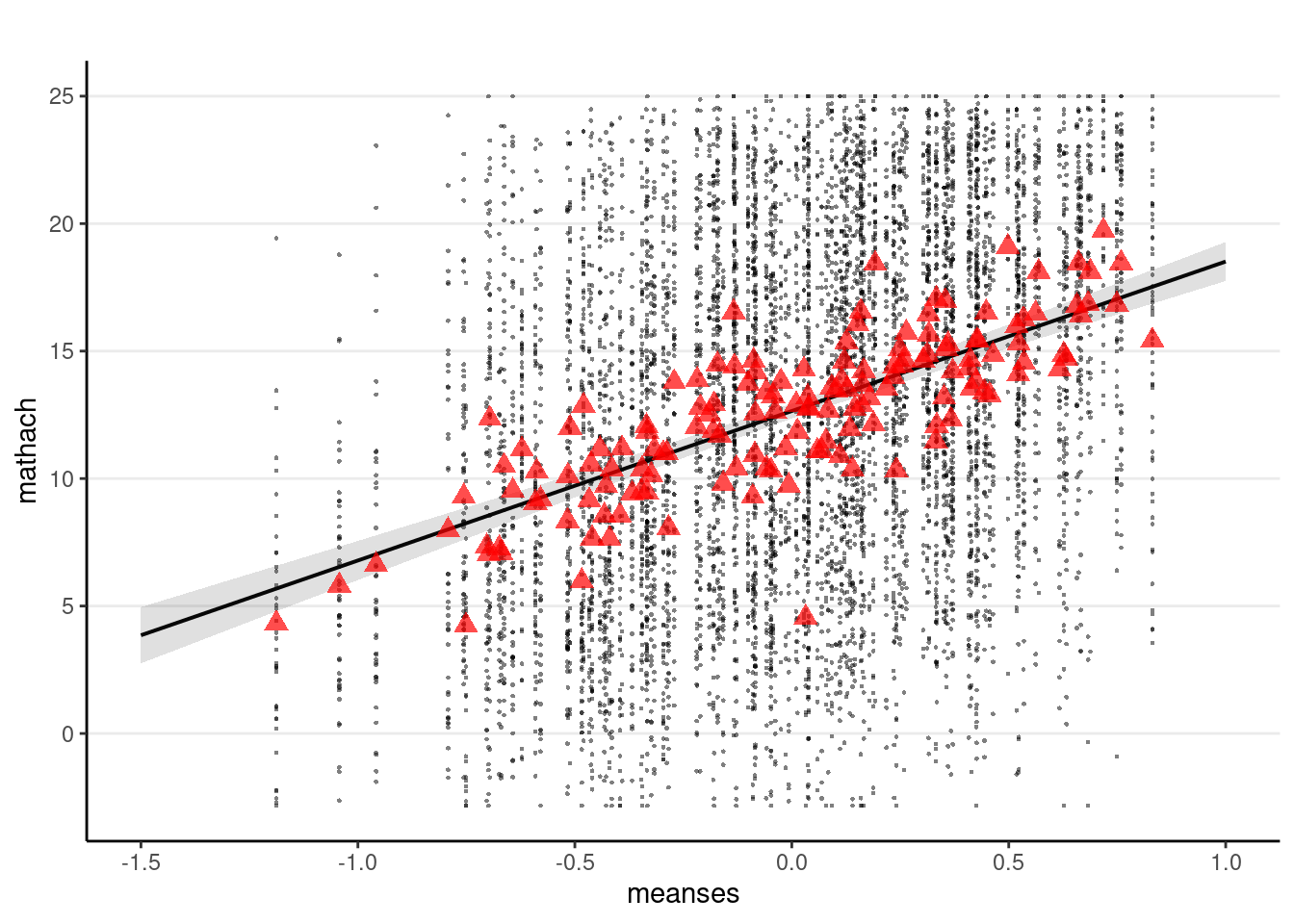
sjPlot::plot_model(m_lv2,
type = "pred", terms = "meanses",
show.data = TRUE, title = "",
dot.size = 0.5
) +
# Add the group means
stat_summary(
data = hsball, aes(x = meanses, y = mathach),
fun = mean, geom = "point",
col = "red",
shape = 17,
# use triangles
size = 3,
alpha = 0.7
)
Proportion of variance predicted
We will use the
# Generally, you should use the marginal R^2 (R2m) for the variance predicted by
# your predictors (`meanses` in this case).
MuMIn::r.squaredGLMM(m_lv2)Warning: 'r.squaredGLMM' now calculates a revised statistic. See the help page. R2m R2c
[1,] 0.1233346 0.1786815An alternative, more comprehensive approach is by Rights & Sterba (2019, Psychological Methods, https://doi.org/10.1037/met0000184), with the r2mlm package
r2mlm::r2mlm(m_lv2)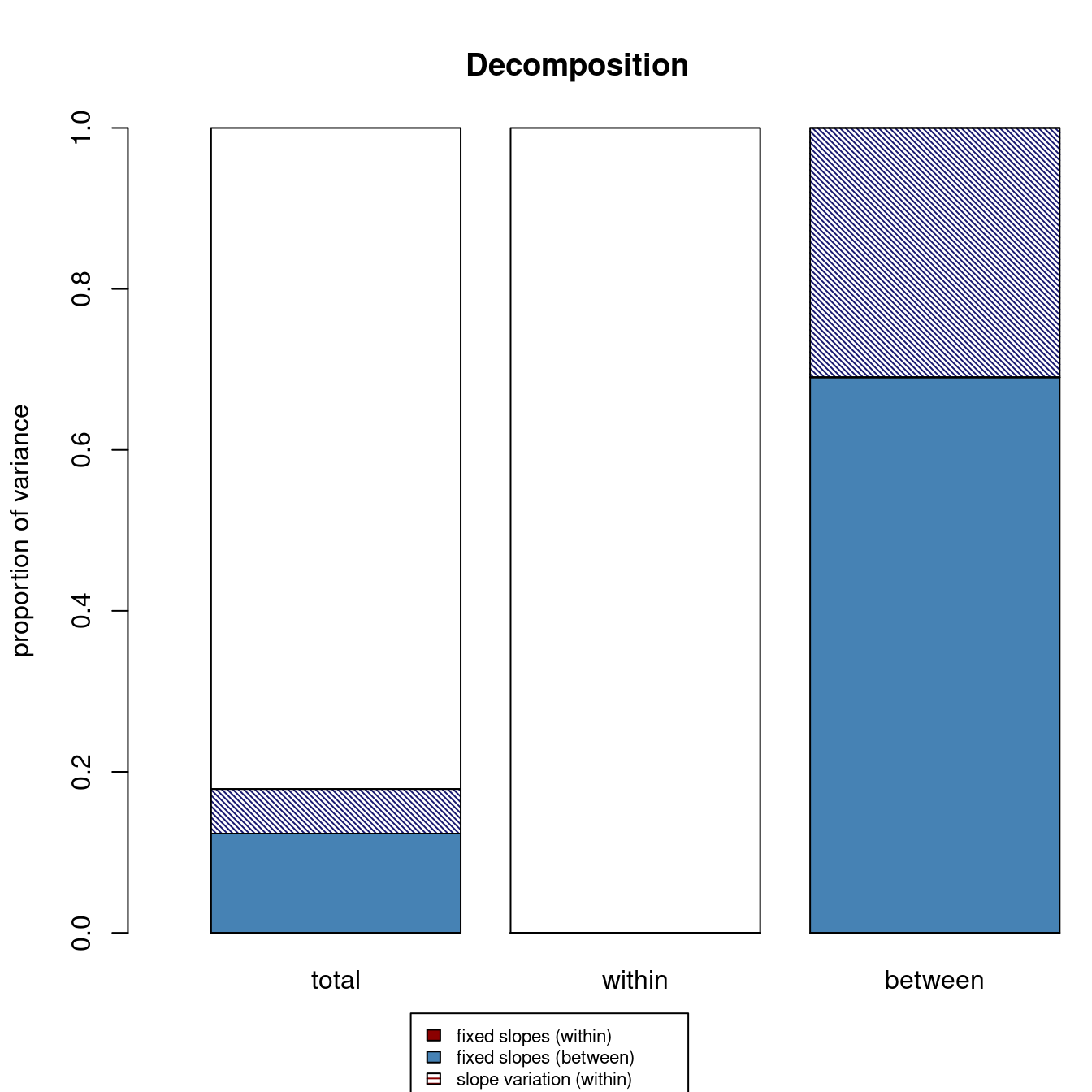
$Decompositions
total within between
fixed, within 0.00000000 0 NA
fixed, between 0.12333464 NA 0.6902487
slope variation 0.00000000 0 NA
mean variation 0.05534682 NA 0.3097513
sigma2 0.82131854 1 NA
$R2s
total within between
f1 0.00000000 0 NA
f2 0.12333464 NA 0.6902487
v 0.00000000 0 NA
m 0.05534682 NA 0.3097513
f 0.12333464 NA NA
fv 0.12333464 0 NA
fvm 0.17868146 NA NANote the fixed, between number in the total column is the same as the one from MuMIn::r.squaredGLMM(). Including school means of SES in the model accounted for about 12% of the total variance of math achievement.
Comparing to OLS regression
Notice that the standard error with regression is only half of that with MLM.
m_lm <- lm(mathach ~ meanses, data = hsball)
msummary(list("MLM" = m_lv2,
"Linear regression" = m_lm))| MLM | Linear regression | |
|---|---|---|
| (Intercept) | 12.649 | 12.713 |
| (0.149) | (0.076) | |
| meanses | 5.864 | 5.717 |
| (0.361) | (0.184) | |
| SD (Intercept id) | 1.624 | |
| SD (Observations) | 6.258 | |
| Num.Obs. | 7185 | 7185 |
| R2 | 0.118 | |
| R2 Adj. | 0.118 | |
| R2 Marg. | 0.123 | |
| R2 Cond. | 0.179 | |
| AIC | 46969.3 | 47202.4 |
| BIC | 46996.8 | 47223.0 |
| ICC | 0.06 | |
| Log.Lik. | −23598.190 | |
| F | 962.329 | |
| RMSE | 6.21 | 6.46 |